the Creative Commons Attribution 4.0 License.
the Creative Commons Attribution 4.0 License.
Bioaerosols as indicators of central Arctic ice nucleating particle sources
Thomas C. J. Hill
Sonia M. Kreidenweis
Paul J. DeMott
Yutaka Tobo
Jessie M. Creamean
The Arctic is warming at a rapid rate, with implications for microbial communities as the ecosystems change. Some microbes and biogenic materials can affect the persistence of long-lived mixed-phase clouds by serving as ice nucleating particles (INPs). The presence of INPs modulates the cloud phase, and long-term measurements are important to elucidate their seasonal sources and to predict future change. The Multidisciplinary drifting Observatory for the Study of Arctic Climate (MOSAiC) expedition in 2019–2020 provided the first year-long measurements of bioaerosols and INPs in the central Arctic, with 3 d filters for amplicon sequencing and cumulative INP concentrations from −5 to −30 °C. Here, we investigated the INP seasonal cycle and its relation to the seasonal cycle of bacteria and eukaryotes. INPs were greatly elevated and compositionally similar in summer, aligning with a greater prevalence of local bioaerosol sources, but, despite this, a diverse mixture of sources (marine and terrestrial) was present all times. A common broader Arctic INP population is hypothesized for much of the year by comparable coincident data collected in Svalbard and a sensitivity of both the INPs and bioaerosols to large-scale events.
- Article
(2306 KB) - Full-text XML
-
Supplement
(1351 KB) - BibTeX
- EndNote
Arctic surface air temperatures are warming 3 or 4 times faster than the rest of the world from Arctic amplification (Rantanen et al., 2022; Zhou et al., 2024). This surface warming affects Arctic ecosystems, including microbial communities. Land effects include greening of tundra vegetation (Berner et al., 2020); thawing of permafrost containing actively transcribing microbes (Wu et al., 2022); and warming of soil, which can alter the frequency of bacterial taxa (Newsham et al., 2022). Ocean effects include increased Atlantic water influx, which could replace cold-adapted bacteria (Carter-Gates et al., 2020); increased sources of dissolved organic matter that could alter bacterial abundance (Nguyen et al., 2022); increased light from sea ice loss, resulting in earlier onset of primary production (Lannuzel et al., 2020); and ocean acidification (Gamberg, 2020).
The effects of changing Arctic ecosystems extend to the atmosphere through microbial emissions from the surface. Some aerosols from these sources can affect clouds by serving as ice nucleating particles (INPs). INPs trigger ice formation at temperatures warmer than homogenous freezing (−38 °C) and have many sources, including bacteria, fungi, organics, and mineral dust (Hill et al., 2018; Kanji et al., 2017; Murray et al., 2012). The lifetime, thickness, and phase of Arctic clouds affect the level of Arctic amplification through impacting the surface energy budget (Tan and Storelvmo, 2019). Arctic mixed-phase clouds occur for about 40 % of the year, and most have temperatures between −25 and −5 °C, a range impacted by many INP sources (Shupe et al., 2006). Modeling studies have shown that the treatment of INPs influences the strength of the cloud phase feedback (Tan et al., 2022).
Some aspects of the Arctic aerosol annual cycle are understood, particularly the transport of anthropogenic emissions from lower latitudes, which typically occurs from January to April and is responsible for the Arctic haze phenomenon. In contrast, the summer aerosol, between June and September, is characterized by increased local influence (Schmale et al., 2021) and tends to be dominated by poorly quantified natural sources.
Some local sources of aerosols, enriched with biogenic INPs, include glacial soil dust and leaf litter (Barr et al., 2023; Conen et al., 2016; Tobo et al., 2019) and marine sources, especially near phytoplankton blooms (Creamean et al., 2019; Hartmann et al., 2021; Wilson et al., 2015). Thermokarst regions contain particularly active, unrepresented potential sources of Arctic INPs (Barry et al., 2023b; Creamean et al., 2020). Recent modeling studies highlighted Arctic dust as a potential important contributor of INPs (Kawai et al., 2023; Shi et al., 2022) as glacial soil dust accounted for nearly all dust INPs in the Arctic lower troposphere between June and November (Kawai et al., 2023).
Long-term Arctic INP measurements reporting the annual cycle have mostly occurred at fixed coastal sites (Pereira Freitas et al., 2023; Sze et al., 2023; Tobo et al., 2024; Wex et al., 2019). They detected seasonality in INP concentrations, with INPs active at warmer temperatures () being in the highest abundance in summer. Higher concentrations of airborne bacterial cells and fungal spores were also found in summer (Abrego et al., 2024; Johansen, 1991; Johansen and Hafsten, 1988; Šantl-Temkiv et al., 2019), with an increase in potential local sources (Jensen et al., 2022). However, in the central Arctic over the pack ice, INP measurements have been limited to specific months (Bigg, 1996; Bigg and Leck, 2001; Hartmann et al., 2021; Porter et al., 2022). The Multidisciplinary drifting Observatory for the Study of Arctic Climate (MOSAiC) campaign on the R/V Polarstern provided the first year of aerosol INP and bioaerosol measurements in the central Arctic (Creamean et al., 2022). To identify contributors, to link source samples to potentially the most active INPs, and as a proxy of air mass origin, we report the first annual cycle of central Arctic aerosol bacteria and eukaryotes.
2.1 Sample collection during the MOSAiC expedition
The MOSAiC expedition took place from October 2019 to September 2020 in the central Arctic aboard the German Research Vessel (R/V) Polarstern, separated into five legs. The vessel drifted passively in ice between 4 October–13 December (leg 1), 13 December–24 February (leg 2), 24 February–16 May (leg 3), 19 June–31 July (leg 4), and 21 August–20 September (leg 5). The other periods were the time when the ship was in transit, but we include collected samples between 27 October 2019–25 September 2020 (Fig. S1 in the Supplement). Overviews of this expedition can be found in Nicolaus et al. (2022), Rabe et al. (2022), and Shupe et al. (2022).
Aerosols for DNA and INP analyses were collected on Polarstern's P deck using filter samplers mounted about 15 m above sea level with the US Department of Energy Atmospheric Radiation Measurement (DOE ARM) AMF2 facility. For DNA analyses, polycarbonate filters (0.4 µm Whatman Nuclepore track-etched hydrophilic membranes) were precleaned by soaking in 10 % H2O2 followed by 0.1 µm filtered deionized (DI) water rinses (Uetake et al., 2020). Polycarbonate filters for INP analyses (0.2 µm Whatman Nuclepore track-etched hydrophilic membranes) were precleaned by brief ultrasonication in methanol followed by 0.1 µm filtered DI water rinses (Barry et al., 2021). Filters for both DNA and INP analyses were precleaned and preloaded in Nalgene units in a laminar flow cabinet at Colorado State University (CSU). Identically cleaned 10 µm polycarbonate filters were loaded underneath the 0.2 and 0.4 µm filters to provide a clean support for the sample filter. Filters were typically collected for 3 d periods, with an average total volume of air filtered of 139 500 standard liters (sL: 0 °C; 1013.25 mb) for DNA filters and 88 800 sL for INP filters.
Samples of seawater, sea ice, snow, melt pond water, and open lead ice were collected and used to identify potential local sources of biological aerosols. Source sample metadata, including types, collection dates and times, latitudes and longitudes, and depths, are provided in Table S1 in the Supplement. Seawater includes samples from Polarstern's flow-through seawater tap system (FT) collected at 11 m depth and from a CTD (conductivity, temperature, depth) rosette at 4–7 depths within the upper 400 m. Sea ice cores were collected using a Kovacs II coring system. Ice cores were sectioned into 5–10 cm segments, melted, and then diluted with 0.22 µm filtered seawater. Snow samples were collected from the surface, middle, and bottom of the snow pits. Melt pond and newly formed lead ice samples were collected mainly during the summer months. Protocols are further described in Nicolaus et al. (2022) and Rabe et al. (2022). All filters and samples were stored at −20 °C for the duration of the campaign, during transport, and at CSU until analysis.
2.2 DNA sample analysis
We processed aerosol filters and source samples for the 16S rRNA gene (bacteria). A subset of aerosol filters and source samples were processed for ITS (fungi), and aerosol filters for the 18S rRNA gene were processed for eukaryotic composition. These samples were processed similarly to previous work in our lab (Barry et al., 2023a; Uetake et al., 2020).
A total of 71 aerosol filter samples were extracted. The processing of 64 samples followed the standard extraction protocol: cutting up the filter into pieces, 30 s ultrasonication in 2 mL of nuclease-free water, and concentration with a Microcon DNA Fast Flow Centrifugal Filter. Extraction was done with the DNeasy PowerLyzer Microbial Kit (Qiagen).
A different extraction protocol was employed for 79 source samples and 7 additional aerosol filters processed later. Ice, seawater, melt pond, flow through, and snowmelt filters were pre-processed identically by thawing the samples and filtering approximately 30 mL through a Sterivex (Millipore) 0.22 µm pore filtering unit (∼15 mL for snow). The Sterivex was separated with a PVC pipe cutter (Cruaud et al., 2017), and the filter was detached with a sterile scalpel and cut into pieces. For the water and additional aerosol samples, filter pieces were placed directly into the extraction tubes of the DNeasy PowerSoil Pro Kit (Qiagen). This kit and the modified method were chosen to remove the concentration pre-step where losses may occur and to allow extraction from the filter to proceed directly. Extraction for both batches followed their respective Qiagen protocol, with two elutions used in the final step to improve recovery.
All aerosol and source samples were amplified for 16S rRNA. The V4–V5 region was targeted with the 515yF/926pfR primers (Parada et al., 2016), with cycling conditions following Uetake et al. (2020) and the UCP Multiplex PCR master mix (Qiagen). A total of 29 aerosol and 36 source samples were amplified for ITS. The primers followed Walters et al. (2016), and cycling conditions were 95 °C for 2 min; 37 cycles of 95 °C for 30 s, 55 °C for 60 s, and 72 °C for 60 s; and, finally, a 72 °C hold for 5 min. A total of 29 aerosol samples were amplified for 18S rRNA. We used the Euk1391f-EukBr primer pair detailed in the Earth Microbiome Project (2017). Cycling conditions were also adapted from the Earth Microbiome Project: 94 °C for 3 min; 37 cycles of 94 °C for 45 s, 57 °C for 60 s, and 72 °C for 90 s; and, finally, a 72 °C hold for 10 min.
The primers contained Illumina adapters and were purified with AMPure XP (Beckman Coulter) twice: after the first amplification and second amplification that added sample barcodes (IDT for Illumina Nextera DNA UD Indexes). This second PCR step had cycling conditions of 95 °C for 5 min; 12 cycles of 95 °C for 30 s, 60 °C for 30 s, and 72 °C for 30 s; and, finally, a 72 °C hold for 7 min. This step used the AmpliTaq Gold LD DNA Polymerase (Applied Biosystems). After the second purification, samples were quantified with the Quant-iT™ 1X dsDNA Assay Kits (Invitrogen) on an Enspire plate reader to create an equimolar library. This library was sequenced at the CSU Next Generation Sequencing Core with the Illumina MiSeq Reagent Kit v3 (600-cycle). Source samples for ITS were sequenced later, and so the sample pool was prepared at CSU identically to the prior samples but sequenced at RTL Genomics (Lubbock, TX) using the same sequencing kit.
Next, sequences were demultiplexed in the Illumina BaseSpace Sequence Hub before being imported into QIIME2 version 2024.5 for processing (Bolyen et al., 2019). Reads were denoised with DADA2 (Callahan et al., 2016) to create an amplicon sequence variant (ASV) table. For 16S, pre-formatted reference sequence and taxonomy files were based on SILVA 138 (Quast et al., 2012; Robeson et al., 2020). For 18S, reference sequence and taxonomy files were from the PR2 (Protist Ribosomal Reference) database, version 5.0.0 (Guillou et al., 2012; Vaulot et al., 2023). For ITS, reference sequence and taxonomy files were from the UNITE Community database (all eukaryotes), version 10.0 (Nilsson et al., 2019). Taxonomy assignment used the feature classifier plugin in QIIME2 (Bokulich et al., 2018; Pedregosa et al., 2011). For 16S, any non-bacterial reads were removed (mitochondria, chloroplast, archaea).
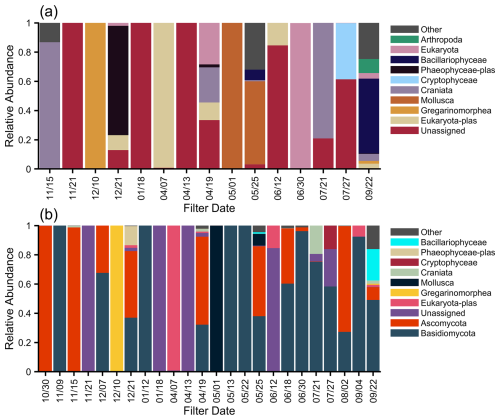
We utilized several field and laboratory controls. Blank aerosol filters were prepared and handled identically to the samples, minus airflow. For water samples, we put 30 mL of nuclease-free water into a 50 mL centrifuge tube and through a Sterivex unit before extraction. Additionally, several extraction and PCR negatives were included. In total, for 16S, three aerosol, four extraction, two Sterivex, and five PCR were done; for ITS, one aerosol, one extraction, one Sterivex, and one PCR were done; and for 18S, one aerosol, two extraction, and two PCR controls were done. An example of the negative- and positive-control taxonomy (from the main aerosol filter sequencing run) is given in Fig. 1. The positive control used was the ZymoBIOMICS® Microbial Community Standard, and this shows that we were able to detect gram negative and positive bacteria in similar percentages.
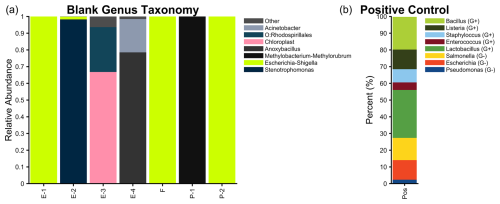
Figure 1(a) The 16S blank relative abundance (genus) for four extraction controls (E-1, E-2, E-3, E-4), one aerosol filter control (F), and two PCR controls (P-1, P-2). (b) The 16S positive-control percentage of sequenced reads.
Since contamination potential is high from the ship and laboratory environment, we did a multi-component blank correction approach for the 16S rRNA aerosol filters. For these, ProkAtlas (Mise and Iwasaki, 2020) was used to assign potential human contamination and is detailed below for environmental use. For blank correction, any ASVs that had more than 50 % attribution from the human environment were excluded. Next, the decontam package prevalence method with a threshold of 0.75 was used (Davis et al., 2018). For the 16S rRNA source, 18S rRNA, and ITS samples, only the decontam step was used, with a threshold of 0.5 for the prevalence method.
For 16S rRNA, 47 aerosol filters and 77 source samples had more than 1000 reads following blank correction and were used for downstream analyses. For ITS aerosol samples, only 10 samples were amplified, but those were amplified well (minimum of 41 287 reads after blank correction). For the ITS source samples, 27 were used for downstream analyses (minimum of 2081 reads after blank correction). For 18S aerosol samples, 24 were used for downstream analyses (minimum of 6268 reads after blank correction).
The analysis methods performed for the 16S rRNA data include sample pooling, source attribution, and alpha diversity. To better represent the bacterial seasonal cycle, the aerosol filters were pooled by month of filter start date for taxonomic plots, using the “mean-ceiling” function to combine the frequencies of identical ASVs. There was only one sample in October (start date of 27 October 2019), which was included with November. For alpha diversity analysis, we rarefied 2269 reads. Source attribution assigned reads to potential environments through ProkAtlas (Mise and Iwasaki, 2020). Categories were collapsed by summing marine and seawater for “marine”; freshwater, glacier, lake water, wastewater, and aquifer for “freshwater”; soil, phyllosphere, rhizosphere, plant, fungus, and terrestrial for “terrestrial”; peat, groundwater, rice paddy, sediment, permafrost, freshwater sediment, and marine sediment for “sediment”; aquatic, salt marsh, hypersaline lake, and estuary for “other water”; gut, insect, insect gut, mouse gut, feces, termite gut, pig gut, rat gut, chicken gut, and bovine gut for “animal association.”
The analysis methods performed for the ITS data included SourceTracker2 (Knights et al., 2011). We rarefied 17 029 reads, which retained 26 source and 10 aerosol samples. We composited the samples into three categories (snow, seawater, and ice and melt pond).
2.3 INP sample analysis
For INP processing of 78 filters, 8 mL of 0.1 µm filtered DI water was added to a filter in a prerinsed 50 mL centrifuge tube to create a suspension and was shaken for 20 min in a Roto-Torque rotater (Cole Parmer). For each, 11-fold dilutions were made (400 µL sample and 4000 µL 0.1 µm filtered DI water) and pipetted out in 32 50 µL aliquots into PCR trays (Optimum Ultra). A 32 50 µL block of 0.1 µm filtered DI water was included with each sample as a negative control. The PCR trays were placed into the aluminum blocks of the CSU Ice Spectrometer (IS) and cooled at 0.33 °C min−1. The current setup of the IS – with the conversion of data into equivalent atmospheric loading (INPs s L−1 of air) and blank corrections – is detailed in DeMott et al. (2018). Four field blanks were transported and processed identically (without airflow) and combined to correct the samples by using an average regression. Blank corrections had virtually no effect on INP concentrations as there was only an average of 11 INPs per blank filter at −25 °C, while sample filters typically had >1000 INPs at this temperature. Thermal and chemical treatments were performed on 26 of the remaining suspensions. These treatments have been used extensively to infer INP composition (Barry et al., 2023a ; Hill et al., 2016; McCluskey et al., 2018; Suski et al., 2018; Testa et al., 2021). Heat treatment at 95 °C removes heat-labile INPs (such as proteins), and hydrogen peroxide (H2O2) digestion at 95 °C removes all organics. Thereby, heat-labile and heat-stable organic INP fractions can be derived, with the remaining fractions being inorganic (presumed mineral).
Weekly INP data obtained at Zeppelin Observatory (78.91° N, 11.89° E) in Svalbard (Pereira Freitas et al., 2023; Tobo et al., 2024) are included for comparison to the data collected on the R/V Polarstern during MOSAiC. The INP data at Zeppelin Observatory were analyzed with the Cryogenic Refrigerator Applied to Freezing Test (CRAFT) system (Tobo, 2016), which uses a cold plate isolated in a clean room. These methods previously compared well with the IS (DeMott et al., 2017). All plots were made with MATLAB ver. R2023b.
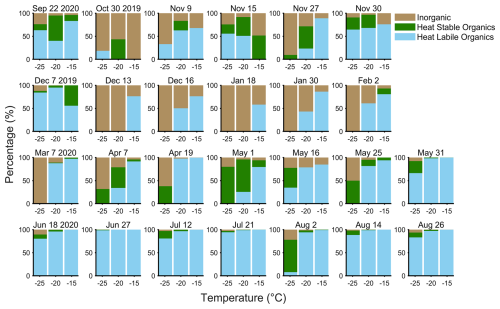
Figure 2Seasonal cycle of the INP composition in the temperature regime at −15, −20, and −25 °C as measured on the R/V Polarstern during MOSAiC. Date refers to the aerosol filter start date, with blue denoting the heat-labile organic fraction (from heating to 95 °C); green denotes the heat-stable organic fraction (from digestion with H2O2); and tan is the inorganic (presumed mineral) contribution from the INPs remaining after H2O2 digestion. Additional information on the seasonal cycle of INP abundances can be found in the Supplement (Fig. S2).
3.1 Seasonal cycle of INP composition
First, we present the seasonal cycle of INP composition for MOSAiC, partially presented in the supplement of Creamean et al. (2022) to add context to their size-resolved data. Based upon heating and H2O2 digestion, we subdivided sample INP populations into heat-labile organics (presumably biological), heat-stable organics (e.g., from soil dust or sea spray), and inorganics (presumably mineral) (Fig. 2). The summer samples were compositionally similar as June through August samples had virtually 100 % heat-labile organic INPs active at both −15 and −20 °C and >80 % heat-labile organic INPs at −25 °C (except on 2 August). The 2–5 August filter was marked by lower concentrations and a lower fraction of heat-labile INPs at −25 °C, which may have resulted from an air mass transition sampling air from predominantly over the ocean to over the sea ice (Fig. 6). Inorganic influence was largest during winter and at the coldest temperatures, which broadly aligns with the Arctic haze period during which dust may also be transported from lower latitudes (Schmale et al., 2022). The Arctic haze season during MOSAiC was stronger and peaked earlier than normal (January and February), with a large positive Arctic Oscillation phase, could have contributed the large inorganic INP fractions seen during this time at temperatures colder than −20 °C (Boyer et al., 2023). However, during all seasons, organic INPs can contribute down to at least −25 °C and are therefore present in the range of mixed-phase clouds.
The heat-labile maximum in summer is reflective of enhanced biological productivity, sea ice minimum, glacial retreat, and less terrestrial snow coverage. This finding also agrees with recent work showing heat-labile fractions of over 90 % in summer and of 50 %–85 % in winter at −12 °C at Zeppelin Observatory (Pereira Freitas et al., 2023) and of 100 % above −20 °C near the North Pole in August and September (Porter et al., 2022). Additionally, we compared the difference between 95 °C heating and an enzymatic digestion on a summer sample (Fig. S9) and found them to be comparable.
3.2 Seasonal cycle of bioaerosols
The Arctic annual cycle of bacterial taxa in ambient aerosol indicates a diverse and complex community (Fig. S3). The psychrophile Polaribacter was the most abundant genus overall at 10 % in aggregate and being detected in 5 months, peaking in summer and fall. This taxon has been found in high abundance in seawater samples after spring phytoplankton blooms in the North Sea (Teeling et al., 2016) and in summer aerosol over the Southern Ocean (Uetake et al., 2020). Among the other MOSAiC aerosol top-20 genera, Sphingomonas, Hymenobacter, and Methylobacterium (Fig. S3), all widely distributed in nature, were previously detected in high relative abundances in air from Station Nord, Greenland, between March and June (Tignat-Perrier et al., 2019). Surprisingly, Fig. S4 shows that bioaerosols had almost no overlap with the major taxa identified in the source samples, suggesting that the main bioaerosol sources were not well represented in the source samples and could be non-local.
Next, we attributed each of our aerosol sequences to their most likely environmental origin to obtain their fractional source contribution (Fig. 3). Potential bacterial contributions attributed to freshwater, sediment, and animals were found in similar proportions throughout the year. The aerosol contained the largest marine influence during summer (52 % in August) and considerable terrestrial influence in all months (>15 %). The alpha diversity of the aerosol samples was not significantly (p<0.05) different between seasons (Fig. S5) despite increased variability in the spring and summer, providing further evidence that the bioaerosols were diverse taxonomic mixtures. Marine influence was at a minimum during the winter and spring despite consistent freshwater contributions, and, when combined with persistent terrestrial influence, this suggests that the bacteria came from longer-range sources as Arctic freshwater sources are frozen during this time. The seasonal cycle of the Arctic haze corresponded to the occurrence of a diverse population of bioaerosols that were most likely from longer-range transport, with a lower population of marine taxa.
Figure 3Potential bacterial source attribution (percentage) based on typical habitat for the pooled aerosol samples as a function of month, combined from ProkAtlas (Mise and Iwasaki, 2020).
The eukaryotic aerosol annual cycle also revealed complex seasonality (Fig. S6). Source tracking of the ITS data (Fig. 4) identified some influence of local sources, especially in the ice and melt pond category during mid-summer. Snow also had ASVs detected in four aerosol samples. The ASVs common to the aerosol and ice and melt pond samples may be attributed to Cryolevonia, which has been isolated from permafrost in the Alps and sea ice (De Garcia et al., 2020; Pontes et al., 2020). These ASVs were previously assigned to this genera in the UNITE 9.0 database but, with 10.0, are only resolved to the class Microbotryomycetes, which contains Cryolevonia (Fig. S6). The sequences common to both the aerosol and local sources may have also originated from more distant Arctic sources, such as an amalgamation of melt ponds (maximum coverage on 30 June: Wang et al., 2024) or re-suspension from previous deposition as the ice near the Polarstern contained Siberian sediment (Krumpen et al., 2020). Nonetheless, sequences in the source and aerosol samples indicate that these categories have the potential to contribute to the bioaerosol and heat-labile INPs.
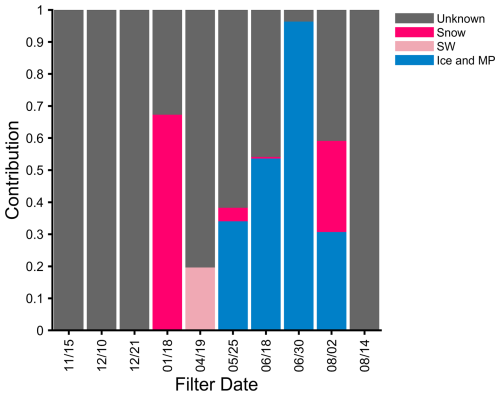
Figure 4ITS source tracking analysis for 10 aerosol samples, organized by month of the campaign (November 2019–August 2020). The listed dates refer to the start date of the filter. A total of 28 source samples were included and composited into three categories: ice and melt pond (MP: blue), seawater (SW: light pink), and snow (dark pink).
The 18S rRNA gene results show that the eukaryotic bioaerosol was dominated by fungi, primarily Basidiomycota and Ascomycota (Fig. 5). Excluding fungi, Bacillariophyceae (diatoms) and Mollusca (mollusks) were observed in multiple spring and summer samples, consistently with increased marine bacterial taxa. Previously, continental work showed a higher normalized species richness of Ascomycota in the winter and spring (Fröhlich-Nowoisky et al., 2009) and at greater relative proportions in marine and/or coastal air (Fröhlich-Nowoisky et al., 2012) due to the smaller spore size of Ascomycota. We generally found increased relative abundance of Basidiomycota in summer; however, the seasonal trends are somewhat ambiguous.
3.3 Investigating potential INP origins through bioaerosol linkage
To investigate bioaerosol and heat-labile INP air mass origins, INPs at −15 °C and biological tracers were plotted with air mass resident time percentages along 5 d HYSPLIT back-trajectories only using points (Fig. 6). This yields the percentage of time spent over the indicated surface type (Creamean et al., 2022). Ice is defined as being greater than 85 % sea ice concentration (SIC), marginal ice zone (MIZ) is 15 %–85 % SIC, ocean is <15 % SIC, and then there is land. Based upon their presence in source samples, we assume that the ASVs were resolved to Microbotryomycetes as a tracer for local and/or regional Arctic sources; Polaribacter, a cold-dwelling marine bacteria, was a tracer for local and/or regional marine air masses; and 18S fungal presence was a general terrestrial marker. Although marine fungi are present in the Arctic (e.g., Hassett et al., 2019), their isolation sources from the Basic Local Alignment Search Tool tool in Geneious Prime 2023.2.1 indicate that the overwhelming majority of our 18S fungal ASVs were terrestrial. Transport of fungal spores from terrestrial sources in summer has been hypothesized, with enhanced concentrations of fluorescent aerosol particles being ascribed to emissions from the tundra (Pereira Freitas et al., 2023; Perring et al., 2023).
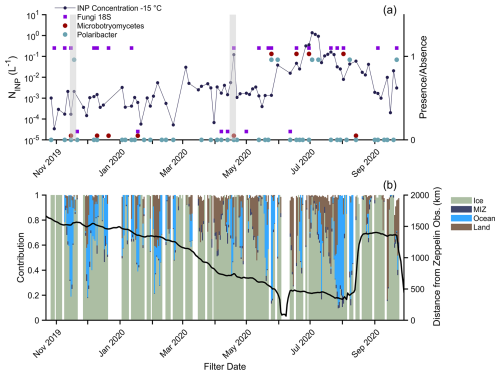
Figure 6(a) INP concentration at −15 °C and corresponding presence (plotted around 1) or absence (plotted around 0) of one fungal taxon (Microbotryomycetes: maroon) and one bacterial taxon (Polaribacter: blue). The presence or absence of fungi is also indicated with purple squares (from the 18S data). (b) Percent contribution to 5 d<500 m back-trajectory and corresponding position of the ship. Ice (green) refers to a sea ice concentration (SIC) greater than 85 %, MIZ (dark blue) is the marginal ice zone and refers to 15 %–85 % SIC, ocean (blue) refers to the ice-free ocean at <15 % SIC, and then there is land (brown). Gray boxes indicate the November storm (16.–20 November 2019) and the April warm air mass intrusion (15–21 April 2020).
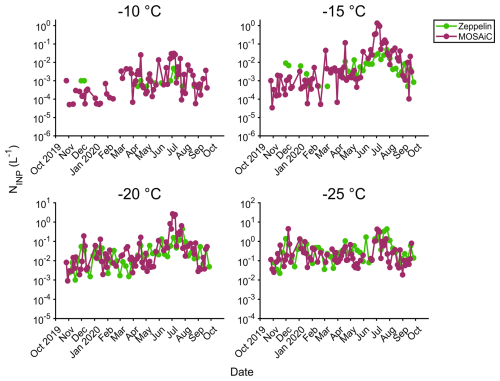
Figure 7INP concentration time series during the MOSAiC campaign at −10, −15, −20, and -25 °C. CRAFT (green) refers to data obtained at Zeppelin Observatory at Svalbard (Pereira Freitas et al., 2023; Tobo et al., 2024). IS (purple) refers to polycarbonate filter samples analyzed with the ice spectrometer.
The seasonal cycle of warm-temperature INPs is evident in Fig. 6, increasing by orders of magnitude from winter to summer. When the highest INP concentrations were observed in summer, the co-occurrence of the regional biological tracers increased. A mixed air mass contribution was usually coincident with the presence of tracers. For example, for the 25–28 May filter, which had daily air mass contribution maxima of 83 % ocean, 77 % ice, and 5 % land, we detected Microbotryomycetes, Polaribacter, and fungi from 18S. Although local sources contributed to the summer aerosol (Fig. 4), these contributions were unlikely to be from a single environment (e.g., solely marine or terrestrial), and regional transport cannot be ruled out.
While INPs followed clear seasonality, their concentrations were affected by periodic large-scale events. Warm air mass intrusions (WAMIs) from cyclones during 16–20 November 2019 and 15–21 April 2020 are shown with gray shading (Fig. 6). The 18S fungi and Polaribacter – but not Microbotryomycetes – were detected during the November storm (Rinke et al., 2021), indicating a mixture of aerosol sources. Back-trajectories indicated a shift from air being predominantly over the ocean on 16 November to being predominantly over the ice on 17–20 November. Polaribacter was not detected until the 18–21 November filter (not sequenced for fungi), and so this air mass transition could be responsible for different bioaerosol populations. Concurrently, INPs at −15 °C increased by an order of magnitude from , with over a 6 °C warmer freezing-onset temperature. After 18–21 November, Polaribacter was not detected again until May 2020, and higher INP concentrations at −15 °C () were not detected until January.
During the 15–21 April WAMI (Dada et al., 2022), the INPs were greatly elevated compared to the periods before and after, reaching 0.12 L−1 at −15 °C. Fungi were only detected in the sample during the storm and not the flanking periods. The air mass compositions were variable, with maxima of 100 % ice, 64 % ice-free ocean, 33 % land, and 8 % MIZ over the event, indicating combined terrestrial and marine influences.
3.4 The regional nature of Arctic INPs
The concentrations and seasonal cycle of INPs observed during MOSAiC agreed well with measurements at Zeppelin Observatory (474 m a.s.l.) in Svalbard (Fig. 7; Pereira Freitas et al., 2023; Tobo et al., 2024), suggesting a commonality of INP concentrations in this Arctic region. Some differences were expected with averaging times: the Zeppelin filters were collected over 1 week and might miss short-term variations. Mean concentrations at −15 °C differed between the sites by only a factor of around 2 in the fall, winter, and spring, but, at MOSAiC, these were as much as an order of magnitude higher in late June and early July (up to 1.4 L−1 at −15 °C), when the Polarstern was an average of 450 km away from Svalbard (Fig. 6). This enhancement could indicate local influence, which agrees with the higher ice and melt pond source attribution of ITS data during this time (Figs. 4 and 5). For activation temperatures of −25 °C, which can be less influenced by biogenic sources (Fig. 2), concentrations were more similar over the year. We note that Fig. 7 indicates much higher INP concentrations than total size-resolved measurements in Creamean et al. (2022): see Sect. S1 and Figs. S7 and S8.
The bioaerosol annual cycle in the central Arctic had a highly variable bacterial and fungal composition. The diversity of taxa and source attribution using 16S and ITS aerosol suggested a mixture of bioaerosols from local and distant sources. Long-range transport episodes were clearly indicated by mixed taxa and increased INP concentrations during WAMI events.
Throughout the annual cycle and across all temperatures, organic (predominantly heat-labile) INPs constituted large fractions of the INP population. These observations led to the surprising conclusion that biological INPs were present year round and dominated the entire INP temperature spectrum in summer. This latter point was further reinforced by the 100-fold increase in INPs active at −15 °C compared to the concentrations during the rest of the year. The frequency of detection of local Arctic fungal and bacterial tracers also increased in summer, and fungal source tracking identified sea ice and melt ponds as potential sources in addition to fungi likely to be from local terrestrial sources with greater relative abundance of Basidiomycota. These observations pointed to increased local contributions to the bioaerosol and, by inference, to the INP population. However, the presence of likely biological INPs throughout the year was unexpected and may be linked to the presence of fungi throughout most of the year.
INP concentrations were largely similar at MOSAiC and Zeppelin Observatory (nearly identical below −20 °C) despite being separated by 300–1600 km (mean=1000 km) horizontally and 500 m vertically. This suggests that these sites sampled a regional-scale INP population arising from mixed sources throughout the year and unlikely to be from point sources proximate to either location, as concluded from the bioaerosol data. Warm-temperature INPs also increased at both sites in summer in response to increased marine biological activity and strong terrestrial sources from decreased snow cover.
In general, these findings are relevant for Arctic mixed-phase clouds as they exist between −25 and −5 °C and are present throughout the year (greatest fractions in spring and fall). The INPs active at temperatures that were observed during MOSAiC in spring and fall had relatively low number concentrations compared with those in summer. As the Arctic warms, the enhanced INP concentrations associated with summertime biological activity could expand into late spring and early fall, changing the annual cycle of INPs and bioaerosols and potentially impacting cloud properties. These data provide an important baseline for evaluating how interannual variability and longer-term trends in Arctic climate manifest in the composition and loading of airborne microorganisms, INP concentrations, and cloud properties.
The DNA data can be accessed at NCBI under accession number PRJNA1086479. The INP data for MOSAiC are published at https://doi.org/10.5439/1804484, and the data for the Zeppelin Observatory in the Arctic Data archive System (ADS) can be found at https://ads.nipr.ac.jp/data/meta/A20230821-002 (Tobo, 2023).
The supplement related to this article is available online at https://doi.org/10.5194/acp-25-11919-2025-supplement.
JMC, SMK, PJDM, and TCJH conceptualized the sampling campaign. KRB and TCJH processed the samples. KRB performed the sample analysis and wrote the paper with contributions from all of the co-authors.
The contact author has declared that none of the authors has any competing interests.
Publisher's note: Copernicus Publications remains neutral with regard to jurisdictional claims made in the text, published maps, institutional affiliations, or any other geographical representation in this paper. While Copernicus Publications makes every effort to include appropriate place names, the final responsibility lies with the authors.
This work was carried out and the data used in this paper were produced as part of the Multidisciplinary drifting Observatory for the Study of Arctic Climate (project no. MOSAiC20192020). The authors would like to thank all of the persons involved in the expedition of the R/V Polarstern during MOSAiC in 2019–2020 (project ID AWI_PS122_00). An extended MOSAiC acknowledgement is given in Nixdorf et al. (2021). Special thanks are given to Jeff Bowman, Emelia Chamberlain, Julia Schmale, Tuija Jokinen, Matthew Shupe, Christian Pilz, Zoe Brasseur, and Tiia Lauila for collecting the source samples and to the staff of the Norwegian Polar Institute for their assistance with year-round measurements at the Zeppelin Observatory. Additionally, we greatly thank the two anonymous reviewers, whose feedback strengthened the paper.
This work was supported by the US Department of Energy Atmospheric Radiation Measurement facility (grant no. DE-AC05-76RL01830), the Atmospheric Systems Research program (grant nos. DE-SC0022046 and DE-SC0019745), JSPS KAKENHI (grant nos. JP19H01972 and JP24H00761), the Arctic Challenge for Sustainability II (ArCS II) Project (grant no. JPMXD1420318865), and the Environment Research and Technology Development Fund (grant nos. JPMEERF20172003, JPMEERF20202003, and JPMEERF20232001) of the Environmental Restoration and Conservation Agency of Japan.
This paper was edited by James Allan and reviewed by two anonymous referees.
Abrego, N., Furneaux, B., Hardwick, B., Somervuo, P., Palorinne, I., Aguilar-Trigueros, C. A., Andrew, N. R., Babiy, U. V., Bao, T., Bazzano, G., Bondarchuk, S. N., Bonebrake, T. C., Brennan, G. L., Bret-Harte, S., Bässler, C., Cagnolo, L., Cameron, E. K., Chapurlat, E., Creer, S., D'Acqui, L. P., de Vere, N., Desprez-Loustau, M., Dongmo, M. A. K., Dyrholm, I. B., Jacobsen, Fisher, B. L., Flores de Jesus, M., Gilbert, G. S., Griffith, G. W., Gritsuk, A. A., Gross, A., Grudd, H., Halme, P., Hanna, R., Hansen, J., Holst Hansen, L., Hegbe, A. D. M. T., Hill, S., Hogg, I. D., Hultman, J., Hyde, K. D., Hynson, N. A., Ivanova, N., Karisto, P., Kerdraon, D., Knorre, A., Krisai-Greilhuber, I., Kurhinen, J., Kuzmina, M., Lecomte, N., Lecomte, E., Loaiza, V., Lundin, E., Meire, A., Mešić, A., Miettinen, O., Monkhouse, N., Mortimer, P., Müller, J., Henrik Nilsson, R., Nonti, P. Y. C., Nordén, J., Nordén, B., Norros, V., Paz, C., Pellikka, P., Pereira, D., Petch, G., Pitkänen, J., Popa, F., Potter, C., Purhonen, J., Pätsi, S., Rafiq, A., Raharinjanahary, D., Rakos, N., Rathnayaka, A. R., Raundrup, K., Rebriev, Y. A., Rikkinen, J., Rogers, H. M. K., Rogovsky, A., Rozhkov, Y., Runnel, K., Saarto, A., Savchenko, A., Schlegel, M., Martin Schmidt, N., Seibold, S., Skjøth, C., Stengel, E., Sutyrina, S. V., Syvänperä, I., Tedersoo, L., Timm, J., Tipton, L., Toju, H., Uscka-Perzanowska, M., van der Bank, M., Herman van der Bank, F., Vandenbrink, B., Ventura, S., Vignisson, S. R., Wang, X., Weisser, W. W., Wijesinghe, S. N., Joseph Wright, S., Yang, C., Yorou, N. S., Young, A., Yu, D. W., Zakharov, E. V., Hebert, P. D. N., Roslin, T., and Ovaskainen, O.: Airborne DNA reveals predictable spatial and seasonal dynamics of fungi, Nature, 631, 835–842, https://doi.org/10.1038/s41586-024-07658-9, 2024.
Barr, S. L., Wyld, B., McQuaid, J. B., Neely Iii, R. R., and Murray, B. J.: Southern Alaska as a source of atmospheric mineral dust and ice-nucleating particles, Sci. Adv., 9, eadg3708, https://doi.org/10.1126/sciadv.adg3708, 2023.
Barry, K. R., Hill, T. C. J., Jentzsch, C., Moffett, B. F., Stratmann, F., and DeMott, P. J.: Pragmatic protocols for working cleanly when measuring ice nucleating particles, Atmos. Res., 250, 105419, https://doi.org/10.1016/j.atmosres.2020.105419, 2021.
Barry, K. R., Hill, T. C. J., Moore, K. A., Douglas, T. A., Kreidenweis, S. M., DeMott, P. J., and Creamean, J. M.: Persistence and Potential Atmospheric Ramifications of Ice-Nucleating Particles Released from Thawing Permafrost, Environ. Sci. Technol., 57, 3505–3515, https://doi.org/10.1021/acs.est.2c06530, 2023a.
Barry, K. R., Hill, T. C. J., Nieto-Caballero, M., Douglas, T. A., Kreidenweis, S. M., DeMott, P. J., and Creamean, J. M.: Active thermokarst regions contain rich sources of ice-nucleating particles, Atmos. Chem. Phys., 23, 15783–15793, https://doi.org/10.5194/acp-23-15783-2023, 2023b.
Berner, L. T., Massey, R., Jantz, P., Forbes, B. C., Macias-Fauria, M., Myers-Smith, I., Kumpula, T., Gauthier, G., Andreu-Hayles, L., Gaglioti, B. V., Burns, P., Zetterberg, P., D'Arrigo, R., and Goetz, S. J.: Summer warming explains widespread but not uniform greening in the Arctic tundra biome, Nat. Commun., 11, 4621, https://doi.org/10.1038/s41467-020-18479-5, 2020.
Bigg, E. K.: Ice forming nuclei in the high Arctic, Tellus B, 48, 223–233, https://doi.org/10.1034/j.1600-0889.1996.t01-1-00007.x, 1996.
Bigg, E. K. and Leck, C.: Cloud-active particles over the central Arctic Ocean, J. Geophys. Res.-Atmos., 106, 32155–32166, https://doi.org/10.1029/1999JD901152, 2001.
Bokulich, N. A., Kaehler, B. D., Rideout, J. R., Dillon, M., Bolyen, E., Knight, R., Huttley, G. A., and Gregory Caporaso, J.: Optimizing taxonomic classification of marker-gene amplicon sequences with QIIME 2's q2-feature-classifier plugin, Microbiome, 6, 90, https://doi.org/10.1186/s40168-018-0470-z, 2018.
Bolyen, E., Rideout, J. R., Dillon, M. R., Bokulich, N. A., Abnet, C. C., Al-Ghalith, G. A., Alexander, H., Alm, E. J., Arumugam, M., Asnicar, F., Bai, Y., Bisanz, J. E., Bittinger, K., Brejnrod, A., Brislawn, C. J., Brown, C. T., Callahan, B. J., Caraballo-Rodríguez, A. M., Chase, J., Cope, E. K., Da Silva, R., Diener, C., Dorrestein, P. C., Douglas, G. M., Durall, D. M., Duvallet, C., Edwardson, C. F., Ernst, M., Estaki, M., Fouquier, J., Gauglitz, J. M., Gibbons, S. M., Gibson, D. L., Gonzalez, A., Gorlick, K., Guo, J., Hillmann, B., Holmes, S., Holste, H., Huttenhower, C., Huttley, G. A., Janssen, S., Jarmusch, A. K., Jiang, L., Kaehler, B. D., Kang, K. B., Keefe, C. R., Keim, P., Kelley, S. T., Knights, D., Koester, I., Kosciolek, T., Kreps, J., Langille, M. G. I., Lee, J., Ley, R., Liu, Y., Loftfield, E., Lozupone, C., Maher, M., Marotz, C., Martin, B. D., McDonald, D., McIver, L. J., Melnik, A. V., Metcalf, J. L., Morgan, S. C., Morton, J. T., Naimey, A. T., Navas-Molina, J. A., Nothias, L. F., Orchanian, S. B., Pearson, T., Peoples, S. L., Petras, D., Preuss, M. L., Pruesse, E., Rasmussen, L. B., Rivers, A., Robeson II, M. S., Rosenthal, P., Segata, N., Shaffer, M., Shiffer, A., Sinha, R., Song, S. J., Spear, J. R., Swafford, A. D., Thompson, L. R., Torres, P. J., Trinh, P., Tripathi, A., Turnbaugh, P. J., Ul-Hasan, S., van der Hooft, J. J. J., Vargas, F., Vázquez-Baeza, Y., Vogtmann, E., von Hippel, M., Walters, W., Wan, Y., Wang, M., Warren, J., Weber, K. C., Williamson, C. H. D., Willis, A. D., Xu, Z. Z., Zaneveld, J. R., Zhang, Y., Zhu, Q., Knight, R., and Caporaso, J. G.: Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2, Nat. Biotechnol., 37, 852–857, https://doi.org/10.1038/s41587-019-0209-9, 2019.
Boyer, M., Aliaga, D., Pernov, J. B., Angot, H., Quéléver, L. L. J., Dada, L., Heutte, B., Dall'Osto, M., Beddows, D. C. S., Brasseur, Z., Beck, I., Bucci, S., Duetsch, M., Stohl, A., Laurila, T., Asmi, E., Massling, A., Thomas, D. C., Nøjgaard, J. K., Chan, T., Sharma, S., Tunved, P., Krejci, R., Hansson, H. C., Bianchi, F., Lehtipalo, K., Wiedensohler, A., Weinhold, K., Kulmala, M., Petäjä, T., Sipilä, M., Schmale, J., and Jokinen, T.: A full year of aerosol size distribution data from the central Arctic under an extreme positive Arctic Oscillation: insights from the Multidisciplinary drifting Observatory for the Study of Arctic Climate (MOSAiC) expedition, Atmos. Chem. Phys., 23, 389–415, https://doi.org/10.5194/acp-23-389-2023, 2023.
Callahan, B. J., McMurdie, P. J., Rosen, M. J., Han, A. W., Johnson, A. J. A., and Holmes, S. P.: DADA2: High-resolution sample inference from Illumina amplicon data, Nat. Methods, 13, 581–583, https://doi.org/10.1038/nmeth.3869, 2016.
Carter-Gates, M., Balestreri, C., Thorpe, S. E., Cottier, F., Baylay, A., Bibby, T. S., Moore, C. M., and Schroeder, D. C.: Implications of increasing Atlantic influence for Arctic microbial community structure, Sci. Rep., 10, 19262, https://doi.org/10.1038/s41598-020-76293-x, 2020.
Conen, F., Stopelli, E., and Zimmermann, L.: Clues that decaying leaves enrich Arctic air with ice nucleating particles, Atmos. Environ., 129, 91–94, https://doi.org/10.1016/j.atmosenv.2016.01.027, 2016.
Creamean, J. M., Cross, J. N., Pickart, R., McRaven, L., Lin, P., Pacini, A., Hanlon, R., Schmale, D. G., Ceniceros, J., Aydell, T., Colombi, N., Bolger, E., and DeMott, P. J.: Ice Nucleating Particles Carried From Below a Phytoplankton Bloom to the Arctic Atmosphere, Geophys. Res. Lett., 46, 8572–8581, https://doi.org/10.1029/2019GL083039, 2019.
Creamean, J. M., Hill, T. C. J., DeMott, P. J., Uetake, J., Kreidenweis, S., and Douglas, T. A.: Thawing permafrost: An overlooked source of seeds for Arctic cloud formation, Environ. Res. Lett., 15, 084022, https://doi.org/10.1088/1748-9326/ab87d3, 2020.
Creamean, J. M., Barry, K., Hill, T. C. J., Hume, C., DeMott, P. J., Shupe, M. D., Dahlke, S., Willmes, S., Schmale, J., Beck, I., Hoppe, C. J. M., Fong, A., Chamberlain, E., Bowman, J., Scharien, R., and Persson, O.: Annual cycle observations of aerosols capable of ice formation in central Arctic clouds, Nat. Commun., 13, 3537, https://doi.org/10.1038/s41467-022-31182-x, 2022.
Cruaud, P., Vigneron, A., Fradette, M.-S., Charette, S. J., Rodriguez, M. J., Dorea, C. C., and Culley, A. I.: Open the Sterivex™ casing: An easy and effective way to improve DNA extraction yields: DNA extraction from Sterivex™ filters, Limnol. Oceanogr.: Methods, 15, 1015–1020, https://doi.org/10.1002/lom3.10221, 2017.
Dada, L., Angot, H., Beck, I., Baccarini, A., Quéléver, L. L. J., Boyer, M., Laurila, T., Brasseur, Z., Jozef, G., De Boer, G., Shupe, M. D., Henning, S., Bucci, S., Dütsch, M., Stohl, A., Petäjä, T., Daellenbach, K. R., Jokinen, T., and Schmale, J.: A central arctic extreme aerosol event triggered by a warm air-mass intrusion, Nat. Commun., 13, 5290, https://doi.org/10.1038/s41467-022-32872-2, 2022.
Davis, N. M., Proctor, D. M., Holmes, S. P., Relman, D. A., and Callahan, B. J.: Simple statistical identification and removal of contaminant sequences in marker-gene and metagenomics data, Microbiome, 6, 226, https://doi.org/10.1186/s40168-018-0605-2, 2018.
De Garcia, V., Trochine, A., Uetake, J., Bellora, N., and Libkind, D.: Novel yeast taxa from the cold: Description of Cryolevonia giraudoae sp. nov. and Camptobasidium gelus sp. nov, Int. J. Syst. Evol. Microbiol., 70, 3711–3717, https://doi.org/10.1099/ijsem.0.004223, 2020.
DeMott, P. J., Hill, T. C. J., Petters, M. D., Bertram, A. K., Tobo, Y., Mason, R. H., Suski, K. J., McCluskey, C. S., Levin, E. J. T., Schill, G. P., Boose, Y., Rauker, A. M., Miller, A. J., Zaragoza, J., Rocci, K., Rothfuss, N. E., Taylor, H. P., Hader, J. D., Chou, C., Huffman, J. A., Pöschl, U., Prenni, A. J., and Kreidenweis, S. M.: Comparative measurements of ambient atmospheric concentrations of ice nucleating particles using multiple immersion freezing methods and a continuous flow diffusion chamber, Atmos. Chem. Phys., 17, 11227–11245, https://doi.org/10.5194/acp-17-11227-2017, 2017.
DeMott, P. J., Möhler, O., Cziczo, D. J., Hiranuma, N., Petters, M. D., Petters, S. S., Belosi, F., Bingemer, H. G., Brooks, S. D., Budke, C., Burkert-Kohn, M., Collier, K. N., Danielczok, A., Eppers, O., Felgitsch, L., Garimella, S., Grothe, H., Herenz, P., Hill, T. C. J., Höhler, K., Kanji, Z. A., Kiselev, A., Koop, T., Kristensen, T. B., Krüger, K., Kulkarni, G., Levin, E. J. T., Murray, B. J., Nicosia, A., O'Sullivan, D., Peckhaus, A., Polen, M. J., Price, H. C., Reicher, N., Rothenberg, D. A., Rudich, Y., Santachiara, G., Schiebel, T., Schrod, J., Seifried, T. M., Stratmann, F., Sullivan, R. C., Suski, K. J., Szakáll, M., Taylor, H. P., Ullrich, R., Vergara-Temprado, J., Wagner, R., Whale, T. F., Weber, D., Welti, A., Wilson, T. W., Wolf, M. J., and Zenker, J.: The Fifth International Workshop on Ice Nucleation phase 2 (FIN-02): laboratory intercomparison of ice nucleation measurements, Atmos. Meas. Tech., 11, 6231–6257, https://doi.org/10.5194/amt-11-6231-2018, 2018.
Fröhlich-Nowoisky, J., Pickersgill, D. A., Després, V. R., and Pöschl, U.: High diversity of fungi in air particulate matter, Proc. Natl. Acad. Sci. USA, 106, 12814–12819, https://doi.org/10.1073/pnas.0811003106, 2009.
Fröhlich-Nowoisky, J., Burrows, S. M., Xie, Z., Engling, G., Solomon, P. A., Fraser, M. P., Mayol-Bracero, O. L., Artaxo, P., Begerow, D., Conrad, R., Andreae, M. O., Després, V. R., and Pöschl, U.: Biogeography in the air: fungal diversity over land and oceans, Biogeosciences, 9, 1125–1136, https://doi.org/10.5194/bg-9-1125-2012, 2012.
Gamberg, M.: Threats to Arctic Ecosystems, Encyclopedia of the World's Biomes, Elsevier, 532–538, https://doi.org/10.1016/B978-0-12-409548-9.11792-0, 2020.
Guillou, L., Bachar, D., Audic, S., Bass, D., Berney, C., Bittner, L., Boutte, C., Burgaud, G., De Vargas, C., Decelle, J., Del Campo, J., Dolan, J. R., Dunthorn, M., Edvardsen, B., Holzmann, M., Kooistra, W. H. C. F., Lara, E., Le Bescot, N., Logares, R., Mahé, F., Massana, R., Montresor, M., Morard, R., Not, F., Pawlowski, J., Probert, I., Sauvadet, A., Siano, R., Stoeck, T., Vaulot, D., Zimmermann, P., and Christen, R.: The Protist Ribosomal Reference database (PR2): A catalog of unicellular eukaryote Small Sub-Unit rRNA sequences with curated taxonomy, Nucleic Acids Res., 41, D597–D604, https://doi.org/10.1093/nar/gks1160, 2012.
Hartmann, M., Gong, X., Kecorius, S., van Pinxteren, M., Vogl, T., Welti, A., Wex, H., Zeppenfeld, S., Herrmann, H., Wiedensohler, A., and Stratmann, F.: Terrestrial or marine – indications towards the origin of ice-nucleating particles during melt season in the European Arctic up to 83.7° N, Atmos. Chem. Phys., 21, 11613–11636, https://doi.org/10.5194/acp-21-11613-2021, 2021.
Hassett, B. T., Borrego, E. J., Vonnahme, T. R., Rämä, T., Kolomiets, M. V., and Gradinger, R.: Arctic marine fungi: Biomass, functional genes, and putative ecological roles, ISME J., 13, 1484–1496, https://doi.org/10.1038/s41396-019-0368-1, 2019.
Hill, T., Barry, K., DeMott, P., and Creamean, J.: MOSAiC-Colorado State University Ice Spectrometer, 2020, U.S. Department of Energy’s Atmospheric Radiation Measurement (ARM) [data set], https://doi.org/10.5439/1804484, 2021.
Hill, T. C. J., DeMott, P. J., Tobo, Y., Fröhlich-Nowoisky, J., Moffett, B. F., Franc, G. D., and Kreidenweis, S. M.: Sources of organic ice nucleating particles in soils, Atmos. Chem. Phys., 16, 7195–7211, https://doi.org/10.5194/acp-16-7195-2016, 2016.
Hill, T. C. J., DeMott, P. J., Conen, F., and Möhler, O.: Impacts of Bioaerosols on Atmospheric Ice Nucleation Processes, in: Microbiology of Aerosols, edited by: Delort, A.-M. and Amato, P., 1st edn., John Wiley and Sons, 197–219, 2018.
Jensen, L. Z., Glasius, M., Gryning, S.-E., Massling, A., Finster, K., and Šantl-Temkiv, T.: Seasonal Variation of the Atmospheric Bacterial Community in the Greenlandic High Arctic Is Influenced by Weather Events and Local and Distant Sources, Front. Microbiol., 13, 909980, https://doi.org/10.3389/fmicb.2022.909980, 2022.
Johansen, S.: Airborne pollen and spores on the Arctic island of Jan Mayen, Grana, 30, 373–379, https://doi.org/10.1080/00173139109431993, 1991.
Johansen, S. and Hafsten, U.: Airborne pollen and spore registrations at Ny-Ålesund, Svalbard, summer 1986, Polar Res., 6, 11–17, https://doi.org/10.3402/polar.v6i1.6842, 1988.
Kanji, Z. A., Ladino, L. A., Wex, H., Boose, Y., Burkert-Kohn, M., Cziczo, D. J., and Krämer, M.: Overview of Ice Nucleating Particles, Meteorol. Monogr., 58, 1.1–1.33, https://doi.org/10.1175/AMSMONOGRAPHS-D-16-0006.1, 2017.
Kawai, K., Matsui, H., and Tobo, Y.: Dominant Role of Arctic Dust With High Ice Nucleating Ability in the Arctic Lower Troposphere, Geophys. Res. Lett., 50, e2022GL102470, https://doi.org/10.1029/2022GL102470, 2023.
Knights, D., Kuczynski, J., Charlson, E. S., Zaneveld, J., Mozer, M. C., Collman, R. G., Bushman, F. D., Knight, R., and Kelley, S. T.: Bayesian community-wide culture-independent microbial source tracking, Nat. Methods, 8, 761–763, https://doi.org/10.1038/nmeth.1650, 2011.
Krumpen, T., Birrien, F., Kauker, F., Rackow, T., von Albedyll, L., Angelopoulos, M., Belter, H. J., Bessonov, V., Damm, E., Dethloff, K., Haapala, J., Haas, C., Harris, C., Hendricks, S., Hoelemann, J., Hoppmann, M., Kaleschke, L., Karcher, M., Kolabutin, N., Lei, R., Lenz, J., Morgenstern, A., Nicolaus, M., Nixdorf, U., Petrovsky, T., Rabe, B., Rabenstein, L., Rex, M., Ricker, R., Rohde, J., Shimanchuk, E., Singha, S., Smolyanitsky, V., Sokolov, V., Stanton, T., Timofeeva, A., Tsamados, M., and Watkins, D.: The MOSAiC ice floe: sediment-laden survivor from the Siberian shelf, The Cryosphere, 14, 2173–2187, https://doi.org/10.5194/tc-14-2173-2020, 2020.
Lannuzel, D., Tedesco, L., Van Leeuwe, M., Campbell, K., Flores, H., Delille, B., Miller, L., Stefels, J., Assmy, P., Bowman, J., Brown, K., Castellani, G., Chierici, M., Crabeck, O., Damm, E., Else, B., Fransson, A., Fripiat, F., Geilfus, N.-X., Jacques, C., Jones, E., Kaartokallio, H., Kotovitch, M., Meiners, K., Moreau, S., Nomura, D., Peeken, I., Rintala, J.-M., Steiner, N., Tison, J.-L., Vancoppenolle, M., Van der Linden, F., Vichi, M., and Wongpan, P.: The future of Arctic sea-ice biogeochemistry and ice-associated ecosystems, Nat. Clim. Change., 10, 983–992, https://doi.org/10.1038/s41558-020-00940-4, 2020.
McCluskey, C. S., Ovadnevaite, J., Rinaldi, M., Atkinson, J., Belosi, F., Ceburnis, D., Marullo, S., Hill, T. C. J., Lohmann, U., Kanji, Z. A., O'Dowd, C., Kreidenweis, S. M., and DeMott, P. J.: Marine and Terrestrial Organic Ice-Nucleating Particles in Pristine Marine to Continentally Influenced Northeast Atlantic Air Masses, J. Geophys. Res.-Atmos., 123, 6196–6212, https://doi.org/10.1029/2017JD028033, 2018.
Mise, K. and Iwasaki, W.: Environmental Atlas of Prokaryotes Enables Powerful and Intuitive Habitat-Based Analysis of Community Structures, iScience, 23, 101624, https://doi.org/10.1016/j.isci.2020.101624, 2020.
Murray, B. J., O'Sullivan, D., Atkinson, J. D., and Webb, M. E.: Ice nucleation by particles immersed in supercooled cloud droplets, Chem. Soc. Rev., 41, 6519, https://doi.org/10.1039/c2cs35200a, 2012.
Newsham, K. K., Danielsen, B. K., Biersma, E. M., Elberling, B., Hillyard, G., Kumari, P., Priemé, A., Woo, C., and Yamamoto, N.: Rapid Response to Experimental Warming of a Microbial Community Inhabiting High Arctic Patterned Ground Soil, Biology, 11, 1819, https://doi.org/10.3390/biology11121819, 2022.
Nguyen, H. T., Lee, Y. M., Hong, J. K., Hong, S., Chen, M., and Hur, J.: Climate warming-driven changes in the flux of dissolved organic matter and its effects on bacterial communities in the Arctic Ocean: A review, Front. Mar. Sci., 9, 968583, https://doi.org/10.3389/fmars.2022.968583, 2022.
Nicolaus, M., Perovich, D. K., Spreen, G., Granskog, M. A., Von Albedyll, L., Angelopoulos, M., Anhaus, P., Arndt, S., Belter, H. J., Bessonov, V., Birnbaum, G., Brauchle, J., Calmer, R., Cardellach, E., Cheng, B., Clemens-Sewall, D., Dadic, R., Damm, E., De Boer, G., Demir, O., Dethloff, K., Divine, D. V., Fong, A. A., Fons, S., Frey, M. M., Fuchs, N., Gabarró, C., Gerland, S., Goessling, H. F., Gradinger, R., Haapala, J., Haas, C., Hamilton, J., Hannula, H.-R., Hendricks, S., Herber, A., Heuzé, C., Hoppmann, M., Høyland, K. V., Huntemann, M., Hutchings, J. K., Hwang, B., Itkin, P., Jacobi, H.-W., Jaggi, M., Jutila, A., Kaleschke, L., Katlein, C., Kotabutin, N., Krampe, D., Kristensen, S. S., Krumpen, T., Kurtz, N., Lampert, A., Lange, B. A., Lei, R., Light, B., Linhardt, F., Liston, G. E., Loose, B., Macfarlane, A. R., Mahmud, M., Matero, I. O., Maus, S., Morgenstern, A., Naderpour, R., Nandan, V., Niubom, A., Oggier, M., Oppelt, N., Pätzold, F., Perron, C., Petrovsky, T., Pirazzini, R., Polashenski, C., Rabe, B., Raphael, I. A., Regnery, J., Rex, M., Ricker, R., Riemann-Campe, K., Rinke, A., Rohde, J., Salganik, E., Scharien, R. K., Schiller, M., Schneebeli, M., Semmling, M., Shimanchuk, E., Shupe, M. D., Smith, M. M., Smolyanitsky, V., Sokolov, V., Stanton, T., Stroeve, J., Thielke, L., Timofeeva, A., Tonboe, R. T., Tavri, A., Tsamados, M., Wagner, D. N., Watkins, D., Webster, M., and Wendisch, M.: Overview of the MOSAiC expedition: Snow and sea ice, Elem. Sci. Anth., 10, 000046, https://doi.org/10.1525/elementa.2021.000046, 2022.
Nilsson, R. H., Larsson, K.-H., Taylor, A. F. S., Bengtsson-Palme, J., Jeppesen, T. S., Schigel, D., Kennedy, P., Picard, K., Glöckner, F. O., Tedersoo, L., Saar, I., Kõljalg, U., and Abarenkov, K.: The UNITE database for molecular identification of fungi: Handling dark taxa and parallel taxonomic classifications, Nucleic Acids Res., 47, D259–D264, https://doi.org/10.1093/nar/gky1022, 2019.
Nixdorf, U., Dethloff, K., Rex, M., Shupe, M., Sommerfeld, A., Perovich, D., Nicolaus, M., Heuzé, C., Rabe, B., Loose, B., Damm, E., Gradinger, R., Fong, A., Maslowski, W., Rinke, A., Kwok, R., Spreen, G., Wendisch, M., Herber, A., Hirsekorn, M., Mohaupt, V., Frickenhaus, S., Immerz, A., Weiss-Tuider, K., König, B., Mengedoht, D., Regnery, J., Gerchow, P., Ransby, D., Krumpen, T., Morgenstern, A., Haas, C., Kanzow, T., Rack, F. R., Saitzev, V., Sokolov, V., Makarov, A., Schwarze, S., Wunderlich, T., Wurr, K., and Boetius, A.: MOSAiC Extended Acknowledgement, 2021.
Parada, A. E., Needham, D. M., and Fuhrman, J. A.: Every base matters: Assessing small subunit rRNA primers for marine microbiomes with mock communities, time series and global field samples: Primers for marine microbiome studies, Environ. Microbiol., 18, 1403–1414, https://doi.org/10.1111/1462-2920.13023, 2016.
Pedregosa, F., Varoquaux, G., Gramfort, A., Michel, V., Thirion, B., Grisel, O., Blondel, M., Prettenhofer, P., Weiss, R., Dubourg, V., Vanderplas, J., Passos, A., and Cournapeau, D.: Scikit-learn: Machine Learning in Python, Machine Learning Research, 12, 2825–2830, 2011.
Pereira Freitas, G., Adachi, K., Conen, F., Heslin-Rees, D., Krejci, R., Tobo, Y., Yttri, K. E., and Zieger, P.: Regionally sourced bioaerosols drive high-temperature ice nucleating particles in the Arctic, Nat. Commun., 14, 5997, https://doi.org/10.1038/s41467-023-41696-7, 2023.
Perring, A. E., Mediavilla, B., Wilbanks, G. D., Churnside, J. H., Marchbanks, R., Lamb, K. D., and Gao, R.: Airborne Bioaerosol Observations Imply a Strong Terrestrial Source in the Summertime Arctic, J. Geophys. Res.-Atmos., 128, e2023JD039165, https://doi.org/10.1029/2023JD039165, 2023.
Pontes, A., Ruethi, J., Frey, B., Aires, A., Thomas, A., Overy, D., Halti, B., Kerr, R., and Sampaio, J. P.: Cryolevonia gen. Nov. and Cryolevonia schafbergensis sp. Nov., a cryophilic yeast from ancient permafrost and melted sea ice, Int. J. Syst. Evol. Microbiol., 70, 2334–2338, https://doi.org/10.1099/ijsem.0.004040, 2020.
Porter, G. C. E., Adams, M. P., Brooks, I. M., Ickes, L., Karlsson, L., Leck, C., Salter, M. E., Schmale, J., Siegel, K., Sikora, S. N. F., Tarn, M. D., Vüllers, J., Wernli, H., Zieger, P., Zinke, J., and Murray, B. J.: Highly Active Ice-Nucleating Particles at the Summer North Pole, J. Geophys. Res.-Atmos., 127, https://doi.org/10.1029/2021JD036059, 2022.
Quast, C., Pruesse, E., Yilmaz, P., Gerken, J., Schweer, T., Yarza, P., Peplies, J., and Glöckner, F. O.: The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools, Nucleic Acids Res., 41, D590–D596, https://doi.org/10.1093/nar/gks1219, 2012.
Rabe, B., Heuzé, C., Regnery, J., Aksenov, Y., Allerholt, J., Athanase, M., Bai, Y., Basque, C., Bauch, D., Baumann, T. M., Chen, D., Cole, S. T., Craw, L., Davies, A., Damm, E., Dethloff, K., Divine, D. V., Doglioni, F., Ebert, F., Fang, Y.-C., Fer, I., Fong, A. A., Gradinger, R., Granskog, M. A., Graupner, R., Haas, C., He, H., He, Y., Hoppmann, M., Janout, M., Kadko, D., Kanzow, T., Karam, S., Kawaguchi, Y., Koenig, Z., Kong, B., Krishfield, R. A., Krumpen, T., Kuhlmey, D., Kuznetsov, I., Lan, M., Laukert, G., Lei, R., Li, T., Torres-Valdés, S., Lin, Li., Lin, Lo., Liu, H., Liu, N., Loose, B., Ma, X., McKay, R., Mallet, M., Mallett, R. D. C., Maslowski, W., Mertens, C., Mohrholz, V., Muilwijk, M., Nicolaus, M., O'Brien, J. K., Perovich, D., Ren, J., Rex, M., Ribeiro, N., Rinke, A., Schaffer, J., Schuffenhauer, I., Schulz, K., Shupe, M., D., Shaw, W., Sokolov, V., Sommerfeld, A., Spreen, G., Stanton, T., Stephens, M., Su, J., Sukhikh, N., Sundfjord, A., Thomisch, K., Tippenhauer, S., Toole, J. M., Vredenborg, M., Walter, M., Wang, H., Wang, L., Wang, Y., Wendisch, M., Zhao, J., Zhou, M., and Zhu, J.: Overview of the MOSAiC expedition: Physical oceanography, Elem. Sci. Anth., 10, 00062, https://doi.org/10.1525/elementa.2021.00062, 2022.
Rantanen, M., Karpechko, A. Yu., Lipponen, A., Nordling, K., Hyvärinen, O., Ruosteenoja, K., Vihma, T., and Laaksonen, A.: The Arctic has warmed nearly four times faster than the globe since 1979, Commun. Earth Environ., 3, 168, https://doi.org/10.1038/s43247-022-00498-3, 2022.
Rinke, A., Cassano, J. J., Cassano, E. N., Jaiser, R., and Handorf, D.: Meteorological conditions during the MOSAiC expedition, Elementa: Sci. Anthropocene, 9, 00023, https://doi.org/10.1525/elementa.2021.00023, 2021.
Robeson, M. S., O'Rourke, D. R., Kaehler, B. D., Ziemski, M., Dillon, M. R., Foster, J. T., and Bokulich, N. A.: RESCRIPt: Reproducible sequence taxonomy reference database management for the masses [Preprint], Bioinformatics, https://doi.org/10.1101/2020.10.05.326504, 2020.
Šantl-Temkiv, T., Lange, R., Beddows, D., Rauter, U., Pilgaard, S., Dall'Osto, M., Gunde-Cimerman, N., Massling, A., and Wex, H.: Biogenic Sources of Ice Nucleating Particles at the High Arctic Site Villum Research Station, Environ Sci. Technol., 53, 10580–10590, https://doi.org/10.1021/acs.est.9b00991, 2019.
Schmale, J., Zieger, P., and Ekman, A. M. L.: Aerosols in current and future Arctic climate, Nat. Clim. Change, 11, 95–105, https://doi.org/10.1038/s41558-020-00969-5, 2021.
Schmale, J., Sharma, S., Decesari, S., Pernov, J., Massling, A., Hansson, H.-C., von Salzen, K., Skov, H., Andrews, E., Quinn, P. K., Upchurch, L. M., Eleftheriadis, K., Traversi, R., Gilardoni, S., Mazzola, M., Laing, J., and Hopke, P.: Pan-Arctic seasonal cycles and long-term trends of aerosol properties from 10 observatories, Atmos. Chem. Phys., 22, 3067–3096, https://doi.org/10.5194/acp-22-3067-2022, 2022.
Shi, Y., Liu, X., Wu, M., Zhao, X., Ke, Z., and Brown, H.: Relative importance of high-latitude local and long-range-transported dust for Arctic ice-nucleating particles and impacts on Arctic mixed-phase clouds, Atmos. Chem. Phys., 22, 2909–2935, https://doi.org/10.5194/acp-22-2909-2022, 2022.
Shupe, M. D., Matrosov, S. Y., and Uttal, T.: Arctic Mixed-Phase Cloud Properties Derived from Surface-Based Sensors at SHEBA, J. Atmos. Sci., 63, 697–711, https://doi.org/10.1175/JAS3659.1, 2006.
Shupe, M. D., Rex, M., Blomquist, B., Persson, P. O. G., Schmale, J., Uttal, T., Althausen, D., Angot, H., Archer, S., Bariteau, L., Beck, I., Bilberry, J., Bucci, S., Buck, C., Boyer, M., Brasseur, Z., Brooks, I. M., Calmer, R., Cassano, J., Castro, V., Chu, D., Costa, D., Cox, C. J., Creamean, J., Crewell, S., Dahlke, S., Damm, E., de Boer, G., Deckelmann, H., Dethloff, K., Dütsch, M., Ebell, K., Ehrlich, A., Ellis, J., Engelmann, R., Fong, A. A., Frey, M. M., Gallagher, M. R., Ganzeveld, L., Gradinger, R., Graeser, J., Greenamyer, V., Griesche, H., Griffiths, S., Hamilton, J., Heinemann, G., Helmig, D., Herber, A., Heuzé, C., Hofer, J., Houchens, T., Howard, D., Inoue, J., Jacobi, H.-W., Jaiser, R., Jokinen, T., Jourdan, O., Jozef, G., King, W., Kirchgaessner, A., Klingebiel, M., Krassovski, M., Krumpen, T., Lampert, A., Landing, W., Laurila, T., Lawrence, D., Lonardi, M., Loose, B., Lüpkes, C., Maahn, M., Macke, A., Maslowski, W., Marsay, C., Maturilli, M., Mech, M., Morris, S., Moser, M., Nicolaus, M., Ortega, P., Osborn, J., Pätzold, F., Perovich, D. K., Petäjä, T., Pilz, C., Pirazzini, R., Posman, K., Powers, H., Pratt, K. A., Preußer, A., Quéléver, L., Radenz, M., Rabe, B., Rinke, A., Sachs, T., Schulz, A., Siebert, H., Silva, T., Solomon, A., Sommerfeld, A., Spreen, G., Stephens, M., Stohl, A., Svensson, G., Uin, J., Viegas, J., Voigt, C., von der Gathen, P., Wehner, B., Welker, J. M., Wendisch, M., Werner, M., Xie, Z., and Yue, F.: Overview of the MOSAiC expedition: Atmosphere, Elem. Sci. Anth., 10, 00060, https://doi.org/10.1525/elementa.2021.00060, 2022.
Suski, K. J., Hill, T. C. J., Levin, E. J. T., Miller, A., DeMott, P. J., and Kreidenweis, S. M.: Agricultural harvesting emissions of ice-nucleating particles, Atmos. Chem. Phys., 18, 13755–13771, https://doi.org/10.5194/acp-18-13755-2018, 2018.
Sze, K. C. H., Wex, H., Hartmann, M., Skov, H., Massling, A., Villanueva, D., and Stratmann, F.: Ice-nucleating particles in northern Greenland: annual cycles, biological contribution and parameterizations, Atmos. Chem. Phys., 23, 4741–4761, https://doi.org/10.5194/acp-23-4741-2023, 2023.
Tan, I. and Storelvmo, T.: Evidence of Strong Contributions From Mixed-Phase Clouds to Arctic Climate Change, Geophys. Res. Lett., 46, 2894–2902, https://doi.org/10.1029/2018GL081871, 2019.
Tan, I., Barahona, D., and Coopman, Q.: Potential Link Between Ice Nucleation and Climate Model Spread in Arctic Amplification, Geophys. Res. Lett., 49, https://doi.org/10.1029/2021GL097373, 2022.
Teeling, H., Fuchs, B. M., Bennke, C. M., Krüger, K., Chafee, M., Kappelmann, L., Reintjes, G., Waldmann, J., Quast, C., Glöckner, F. O., Lucas, J., Wichels, A., Gerdts, G., Wiltshire, K. H., and Amann, R. I.: Recurring patterns in bacterioplankton dynamics during coastal spring algae blooms, eLife, 5, e11888, https://doi.org/10.7554/eLife.11888, 2016.
Testa, B., Hill, T. C. J., Marsden, N. A., Barry, K. R., Hume, C. C., Bian, Q., Uetake, J., Hare, H., Perkins, R. J., Möhler, O., Kreidenweis, S. M., and DeMott, P. J.: Ice Nucleating Particle Connections to Regional Argentinian Land Surface Emissions and Weather During the Cloud, Aerosol, and Complex Terrain Interactions Experiment, J. Geophys. Res.-Atmos., 126, https://doi.org/10.1029/2021JD035186, 2021.
Tignat-Perrier, R., Dommergue, A., Thollot, A., Keuschnig, C., Magand, O., Vogel, T. M., and Larose, C.: Global airborne microbial communities controlled by surrounding landscapes and wind conditions, Sci. Rep., 9, 14441, https://doi.org/10.1038/s41598-019-51073-4, 2019.
Tobo, Y.: An improved approach for measuring immersion freezing in large droplets over a wide temperature range, Sci. Rep., 6, 32930, https://doi.org/10.1038/srep32930, 2016.
Tobo, Y.: Monitoring of atmospheric ice nucleating particle (INP) number concentrations at the Zeppelin Observatory, Svalbard (MOSAiC), 2020, 1.00, Arctic Data archive System (ADS) [data set], https://ads.nipr.ac.jp/dataset/A20230821-002 (last access: 26 September 2025), 2023.
Tobo, Y., Adachi, K., DeMott, P. J., Hill, T. C. J., Hamilton, D. S., Mahowald, N. M., Nagatsuka, N., Ohata, S., Uetake, J., Kondo, Y., and Koike, M.: Glacially sourced dust as a potentially significant source of ice nucleating particles, Nat. Geosci., 12, 253–258, https://doi.org/10.1038/s41561-019-0314-x, 2019.
Tobo, Y., Adachi, K., Kawai, K., Matsui, H., Ohata, S., Oshima, N., Kondo, Y., Hermansen, O., Uchida, M., Inoue, J., and Koike, M.: Surface warming in Svalbard may have led to increases in highly active ice-nucleating particles, Commun. Earth Environ., 5, 516, https://doi.org/10.1038/s43247-024-01677-0, 2024.
Uetake, J., Hill, T. C. J., Moore, K. A., DeMott, P. J., Protat, A., and Kreidenweis, S. M.: Airborne bacteria confirm the pristine nature of the Southern Ocean boundary layer, P. Natl. Acad. Sci. USA, 117, 13275–13282, https://doi.org/10.1073/pnas.2000134117, 2020.
Vaulot, D., Del Campo, J., Burki, F., Jamy, M., Guillou, L., Santoferrara, L., Ganser, M., de Oliveira da Rocha Franco, A., Mertens, K., Gu, H., Hyeon Jang, S., Škaloud, P., Dünn, M., Gross, M., Seliuk, A., Sandin, M., Metz, S., Fiore-Donno, A. M., and Dorrell, R.: PR2 version 5.0.0 [data set], https://doi.org/10.5281/zenodo.7805244, 2023.
Walters, W., Hyde, E. R., Berg-Lyons, D., Ackermann, G., Humphrey, G., Parada, A., Gilbert, J. A., Jansson, J. K., Caporaso, J. G., Fuhrman, J. A., Apprill, A., and Knight, R.: Improved Bacterial 16S rRNA Gene (V4 and V4–5) and Fungal Internal Transcribed Spacer Marker Gene Primers for Microbial Community Surveys, mSystems, 1, e00009-15, https://doi.org/10.1128/mSystems.00009-15, 2016.
Wang, M., Linhardt, F., Lion, V., and Oppelt, N.: Melt Pond Evolution along the MOSAiC Drift: Insights from Remote Sensing and Modeling, Remote Sens., 16, 3748, https://doi.org/10.3390/rs16193748, 2024.
Wex, H., Huang, L., Zhang, W., Hung, H., Traversi, R., Becagli, S., Sheesley, R. J., Moffett, C. E., Barrett, T. E., Bossi, R., Skov, H., Hünerbein, A., Lubitz, J., Löffler, M., Linke, O., Hartmann, M., Herenz, P., and Stratmann, F.: Annual variability of ice-nucleating particle concentrations at different Arctic locations, Atmos. Chem. Phys., 19, 5293–5311, https://doi.org/10.5194/acp-19-5293-2019, 2019.
Wilson, T. W., Ladino, L. A., Alpert, P. A., Breckels, M. N., Brooks, I. M., Browse, J., Burrows, S. M., Carslaw, K. S., Huffman, J. A., Judd, C., Kilthau, W. P., Mason, R. H., McFiggans, G., Miller, L. A., Nájera, J. J., Polishchuk, E., Rae, S., Schiller, C. L., Si, M., Vergara Temprado, J., Whale, T. F., Wong, J. P. S., Wurl, O., Yakobi-Hancock, J. D., Abbatt, J., P. D., Aller, J. Y., Bertram, A. K., Knopf, D. A., and Murray, B. J.: A marine biogenic source of atmospheric ice-nucleating particles, Nature, 525, 234–238, https://doi.org/10.1038/nature14986, 2015.
Wu, R., Trubl, G., Taş, N., and Jansson, J. K.: Permafrost as a potential pathogen reservoir, One Earth, 5, 351–360, https://doi.org/10.1016/j.oneear.2022.03.010, 2022.
Zhou, W., Leung, L. R., and Lu, J.: Steady threefold Arctic amplification of externally forced warming masked by natural variability, Nat. Geosci., 17, 508–515, https://doi.org/10.1038/s41561-024-01441-1, 2024.