the Creative Commons Attribution 4.0 License.
the Creative Commons Attribution 4.0 License.
Effects of pH and light exposure on the survival of bacteria and their ability to biodegrade organic compounds in clouds: implications for microbial activity in acidic cloud water
Yushuo Liu
Chee Kent Lim
Zhiyong Shen
Patrick K. H. Lee
Recent studies have reported that interactions between live bacteria and organic matter can potentially affect the carbon budget in clouds, which has important atmospheric and climate implications. However, bacteria in clouds are subject to a variety of atmospheric stressors, which can adversely affect their survival and energetic metabolism and, consequently, their ability to biodegrade organic compounds. At present, the effects of cloud water pH and solar radiation on bacteria are not well understood. In this study, we investigated how cloud water pH (pH 3 to 6) and exposure to solar radiation impact the survival and energetic metabolism of two Enterobacter bacterial strains that were isolated from ambient air collected in Hong Kong and their ability to biodegrade organic acids. Experiments were conducted using simulated sunlight (wavelength from 320 to 700 nm) and microcosms comprised of artificial cloud water that mimicked the pH and chemical composition of cloud water in Hong Kong, South China. Our results showed that the energetic metabolism and survival of both strains depended on the pH. Low survival rates were observed for both strains at pH<4, regardless of whether the strains were exposed to simulated sunlight. At pH 4 to 5, the energetic metabolism and survival of both strains were negatively impacted only when they were exposed to simulated sunlight. Organic compounds such as lipids and peptides were detected during exposure to simulated sunlight at pH 4 to 5. In contrast, there were minimal effects on the energetic metabolism and the survival of both strains when they were exposed to simulated sunlight at pH>5. The biodegradation of organic acids was found to depend on the presence (or absence) of simulated sunlight and the pH of the artificial cloud water medium. Overall, this study provides new insights into how two common atmospheric stressors, cloud water pH and exposure to solar radiation, can influence the survival and energetic metabolism of bacteria, and consequently the roles that they play in cloud processes.
- Article
(4507 KB) - Full-text XML
-
Supplement
(2488 KB) - BibTeX
- EndNote
Clouds are an important medium for the aqueous-phase formation and transformation of organic and inorganic compounds. In addition to inorganic and organic compounds, clouds contain biological matter including biological debris (e.g., dead cells and cell fragments) and live microorganisms (e.g., bacteria and fungal spores; Bauer et al., 2002; Jaenicke, 2005; Burrows et al., 2009). Live microorganisms are mainly emitted directly into the atmosphere from natural sources (Jaenicke, 2005; Möhler et al., 2007; Burrows et al., 2009; Attard et al., 2012; Hu et al., 2018). Once airborne, they can participate in a variety of atmospheric processes such as cloud formation, precipitation, ice nucleation, and the microbial degradation of atmospheric organics (Amato et al., 2005; Delort et al., 2010; Vaitilingom et al., 2010, 2013; Morris et al., 2014, 2017; Hu et al., 2018; Huang et al., 2021; Zhang et al., 2021). Bacteria are incorporated into clouds through nucleation and scavenging processes (Möhler et al., 2007). So far, only bacterial communities in clouds in some areas (e.g., Puy-de-Dôme in France and Mount Tai in North China) have been extensively investigated. These studies showed that the bacterial communities in clouds are highly complex and diverse and mainly originate from vegetation, soil, and waterbodies (Vaïtilingom et al., 2012; Wei et al., 2017; Zhu et al., 2018). A significant fraction of the bacteria in clouds may be major allergens and/or pathogens that originate mainly from anthropogenic activities, and their concentrations usually increase during air pollution episodes (Wei et al., 2017; Peng et al., 2019). The cell concentrations of bacteria in clouds typically range from about 102 to 105 cells mL−1 (Amato et al., 2005, 2017; Burrows et al., 2009). At present, our knowledge on bacterial communities in clouds is limited to the few areas that have been studied (e.g., Puy-de-Dôme in France and Mount Tai in North China; Amato et al., 2005, 2017; Wei et al., 2017;Péguilhan et al., 2021). Cultural bacteria typically makes up a very small fraction (about 1 %) of the entire bacteria community in clouds (Amato et al., 2005).
Airborne bacteria are comprised of both dead or dormant cells and metabolically active cells. Previous culture-based and culture-independent analyses of bacteria isolated from cloud water have shown that some of these bacteria species are metabolically active (Amato et al., 2007, 2019; Krumins et al., 2014). Previous studies have reported that the degradation of organic compounds as a result of microbiological–chemical interactions between live bacteria and organic matter can play an important role in influencing the carbon budget in clouds, which will have important atmospheric and climate implications (Delort et al., 2010; Vaitilingom et al., 2010, 2013; Ervens and Amato, 2020). Many bacteria species have the enzymes needed to biodegrade organic compounds. Some of the bacteria species isolated from cloud water could biodegrade organic acids, formaldehyde, methanol, phenolic compounds, and amino acids (Ariya et al., 2002; Husárová et al., 2011; Vaïtilingom et al., 2011; Jaber et al., 2020, 2021). However, the bacteria are exposed to a variety of stressors that can negatively impact their survival and microbial activity in clouds. Joly et al. (2015) previously investigated the individual impacts of osmotic shocks, freeze–thaw cycles, and exposure to light and H2O2 on the survival of different bacterial strains in microcosms mimicking cloud water chemical composition at Puy-de-Dôme. Osmotic shocks and freeze–thaw cycles reportedly had the greatest negative impacts on the survival of bacteria, while exposure to light and H2O2 had limited impacts on the survival of bacteria. However, there are other stressors that bacteria in clouds are commonly subjected to beyond the four stressors investigated by Joly et al. (2015). In addition, when combined together, the stressors may have synergistic negative impacts on the survival and microbial activity of bacteria in clouds. The potentially synergistic negative impacts that stressors have on the survival and microbial activity of bacteria in clouds have yet to be investigated. Some bacteria species respond to stressors by releasing organic compounds (e.g., proteins, pigments, and lipids) as a defensive mechanism (Davey and O'toole, 2000; Delort et al., 2010; Flemming and Wingender, 2010; Vaïtilingom et al., 2012; Matulova et al., 2014). When bacteria species cannot withstand the stress, the resulting cellular damage and lysis will lead to the release of biological material. In addition, the ability of bacteria to biodegrade organic compounds in clouds will decrease if their metabolism and survival are negatively impacted.
Cloud water acidity is another stressor that bacteria are subjected to in clouds. There has been limited study on the impact of cloud water pH on the survival and microbial activity of bacteria in clouds. However, some studies have reported that the cloud water pH impacts the diversity and composition of bacterial communities (Amato et al., 2005; Peng et al., 2019). For instance, spore-forming bacteria were abundant in pH 4.9 cloud water at Puy-de-Dôme, while more diverse and higher concentrations of non-spore-forming bacteria were observed in pH 5.8 cloud water (Amato et al., 2005). The pH of cloud water typically lies between 3 and 6 (Pye et al., 2020), with a global mean of around pH 5.2 (Shah et al., 2020). Areas with high inputs of sulfuric acid and/or nitric acid combined with low inputs of ammonia, dust, and sea salt, especially in parts of East Asia, have moderately acidic to highly acidic cloud water (pH<5; Li et al., 2020; Pye et al., 2020; Shah et al., 2020; Qu and Han, 2021). To the best of our knowledge, there have been no studies on how moderately acidic to highly acidic cloud water affects the survival and microbial activity of bacteria. The effects of light exposure on the survival and microbial activity of bacteria are also ambiguous. Some studies reported that exposure to UVA and visible light will lead to the formation of intracellular reactive oxidative species, which can damage important cell components and cause cell death (Anglada et al., 2015). However, exposure to light reportedly did not impact the survival rates of bacterial strains from Pseudomonas syringae, Arthrobacter sp., and Sphingomonas sp. (Joly et al., 2015). While it is possible that exposure to acidic cloud water and light have a synergistic effect on the survival and microbial activity of bacteria, previous laboratory investigations were mainly performed in microcosms with the pH set between 5 and 7 to mimic cloud water in areas that have high inputs of ammonia, dust, and sea salt, such as the Puy-de-Dôme (Vaïtilingom et al., 2011; Joly et al., 2015; Jaber et al., 2021, 2020).
This study investigates how cloud water pH and exposure to solar radiation affect the survival and energetic metabolism of bacteria and their ability to biodegrade organic compounds in clouds. We designed a series of laboratory experiments in microcosms containing artificial cloud water that mimicked the pH and chemical composition of atmospheric cloud water collected at the Tai Mo Shan station in Hong Kong, South China. South China is a region with moderately acidic to highly acidic cloud water due to its higher concentrations of acidic ions (e.g., and ) compared to alkaline ions (e.g., and Ca2+; Li et al., 2020; Qu and Han, 2021). Different pH (pH 3.3 to 5.9) and irradiation (illuminated vs. dark) conditions were employed in the experiments, during which we analyzed the biological material and organic compounds in the artificial cloud water medium at different reaction time points. Since cloud water bacterial isolates from the Tai Mo Shan station are not available, two Enterobacter bacterial strains that were isolated from ambient air in Hong Kong were used as model bacteria in this study. In general, our current knowledge of the diversity and composition of bacteria communities in cloud water in Hong Kong and South China is very limited due to the scarcity of characterization studies conducted in this region. Results from a previous study reported that Enterobacter was one of the bacteria species in cloud water collected at the Nanling Mountain station in South China (Peng et al., 2019). Enterobacter bacteria have been detected in urban aerosols in different parts of the world, including South China (Chen et al., 2012; Després et al., 2012; Ding et al., 2015; Zhou et al., 2018; Prokof'eva et al., 2021). In addition, the enrichment of Enterobacter bacteria in the atmosphere during air pollution episodes has been reported in parts of Asia, America, and Europe (Romano et al., 2019, 2021; Ruiz-Gil et al., 2020). Since organic acids are ubiquitous in clouds (Tsai and Kuo, 2013; Löflund et al., 2002; Sun et al., 2016; Li et al., 2020) and can be biodegraded by most bacteria (Vaitilingom et al., 2010; Vaïtilingom et al., 2011), we chose seven organic acids that are commonly detected in clouds (formic acid, acetic acid, oxalic acid, maleic acid, malonic acid, glutaric acid, and methanesulfonic acid) as model organic compounds for our investigations of how cloud water pH and light exposure affect the ability of bacteria to biodegrade organic compounds in clouds.
2.1 Strain isolation and whole genome sequencing
Two new strains (B0910 and pf0910) belonging to Enterobacter species were isolated by exposing nutrient agar plates to ambient air in an urban environment (22.3360∘ N, 114.1732∘ E) at a height of 50 m above sea level during the summer season (∼22 ∘C) in Hong Kong. The genomes of the two strains were sequenced using a GridION sequencer (Oxford Nanopore Technologies) by following the manufacturer's workflow. Genome assembly and the downstream genomic analyses are described in detail in Sect. S1 in the Supplement. Based on genome comparison, E. hormaechei B0910 is most similar to Enterobacter hormaechei subsp. hoffmannii DSM 14563 (average nucleotide identity (ANI)=98.92) and E. hormaechei pf0910 to Enterobacter hormaechei subsp. steigerwaltii DSM 16691 (ANI=98.73; Fig. S1 in the Supplement). E. hormaechei B0910 has a chromosome (4.69 Mbp) with 4875 coding sequences (CDSs) and a single plasmid (373 Kbp) with 383 CDSs. E. hormaechei pf0910 strain has a chromosome (4.78 Mbp) with 5072 CDSs and two plasmids of 281 Kbp (344 CDSs) and 73 Kbp (79 CDSs).
2.2 General experimental approach
To simulate cloud water conditions in Hong Kong, artificial cloud water containing major organic and inorganic ions in cloud water previously collected at the Tai Mo Shan station (TMS; N, E, 957 m a.s.l.) were used in each experiment. Organic (acetic acid, formic acid, oxalic acid, and pyruvic acid) and inorganic (magnesium chloride, calcium chloride, potassium chloride, sodium chloride, ammonium sulfate, ammonium nitrate, sodium hydroxide, and hydrochloric acid) compounds were used to prepare the artificial cloud water. Experiments were performed using a Rayonet photoreactor (RPR-200; Southern New England Ultraviolet Company). We followed the method employed in previous studies (George et al., 2015; Huang et al., 2018; Misovich et al., 2021; Li et al., 2022) and used eight lamps with outputs centered at different wavelengths to roughly simulate the range of solar radiation wavelengths (320 to 700 nm) inside the photoreactor. Figure S2 shows the resulting photon flux inside the photoreactor. The temperature (25 ∘C) during the experiment was regulated by a fan located at the bottom of the photoreactor.
The two strains were grown in lysogeny broth (LB) at 37 ∘C to stationary phase. The culture was then centrifuged at 6000 rpm (revolutions per minute) for 10 min at 4 ∘C, and the cell pellets were rinsed with artificial cloud water (Table S1 in the Supplement) three times. For investigations of the time evolution in the survival and energetic metabolism of bacteria at different pH under illuminated vs. dark conditions (Sect. 2.2), the cells were resuspended in artificial cloud water to an initial concentration of ∼105 cells per milliliter. For investigations of the biodegradation of organic acids by bacteria at different pH under illuminated vs. dark conditions (Sect. 2.3), the cells were resuspended in artificial cloud water to an initial concentration of ∼106 cells per milliliter. A calibration curve was used to convert between optical density and bacterial cell concentration.
Quartz tubes containing bacterial cells suspended in artificial cloud water (5 mL) were placed on a rotating vial rack in the middle of the photoreactor. The quartz tubes for the dark control experiments were wrapped in aluminum foil and placed inside the photoreactor. The pH of the artificial cloud water did not change significantly during the experiments. Aliquots of the solutions were taken at every hour over 12 h for various offline chemical analyses. A total of 100 µL of the solution was removed at each time point for colony-forming unit (CFU) counts on LB agar at 37 ∘C for 16 h to determine the culturable bacterial cell concentrations, which was used to calculate the bacteria survival rates. A total of 20 µL of the solution was removed at different each point for measurements of adenosine diphosphate/adenosine triphosphate (ADPATP) ratios, using an assay kit (EnzyLight™, BioAssay Systems) and a biolumineter (SpectraMax M2e) to determine changes in the bacteria energetic metabolism. All the experiments and measurements were performed in triplicate.
2.3 Investigations of the survival and energetic metabolism of bacteria at different pH under illuminated vs. dark conditions
Six pH conditions (pH 3.3, 4.3, 4.5, 4.7, 5.2, and 5.9) were chosen for this set of experiments, which were performed under both dark and illuminated conditions. The six pH conditions investigated fall within the range of pH values for cloud water previously measured at Tai Mo Shan (pH 3.0 to 5.9; Li et al., 2020). The pH of the artificial cloud water used to suspend the bacterial cells was adjusted using sodium hydroxide and hydrochloric acid. Table S1 shows the resulting concentrations of organic and inorganic ions in the artificial cloud water used in these experiments, which are similar to those in cloud water collected at Tai Mo Shan by Li et al. (2020).
During some experiments, aliquots of the solutions (10 mL) were taken at time points 0, 2, 4, 8, and 12 h and analyzed by ultra-performance liquid chromatography mass spectrometry (UPLC-MS). Each aliquot of the solution was first passed through a 0.22 µm filter to remove intact bacterial cells. Water-insoluble and water-soluble biological material and organic compounds were then extracted from these filtered solutions using the method described in Sect. S2. In total, 200 µL of the extract were then transferred into glass vial inserts for UPLC-MS analysis. Non-targeted UPLC-MS analysis was performed using an ultrahigh-performance liquid chromatography system (ExionLC AD system; Sciex) coupled to a high-resolution quadrupole-time-of-flight mass spectrometer (TripleTOF 6600 system, Sciex) equipped with electrospray ionization (ESI). Chromatographic separation was performed on a Kinetex HILIC LC column (100×2.1 mm, 2.6 µm, 100 Å; Phenomenex) using a positive ESI mode. Since very low signals were obtained for a negative ESI mode, we did not use it for our analysis. Details about the UPLC-MS operation, data processing, and statistical analysis can be found in Sect. S3.
2.4 Investigations of the biodegradation of organic acids at different pH under illuminated vs. dark conditions
The biodegradation of seven organic acids (formic acid, acetic acid, oxalic acid, maleic acid, malonic acid, glutaric acid, and methanesulfonic acid (MSA)) that were mixed together were measured at pH 4.3 and pH 5.9 under both dark and illuminated conditions. The concentrations for each of the aforementioned organic acids in cloud water and rainwater typically fall within the range of 1 to 10 µM (Tsai and Kuo, 2013; Löflund et al., 2002; Sun et al., 2016; Li et al., 2020). Due to the detection limits of the IC system used to measure the organic acids, the concentration for each organic acid was set to 50 µM (Table S2), which is around 10 times higher than the concentrations typically measured in cloud water. The concentrations of inorganic ions in the artificial cloud water were also increased by 10 times. Vaitilingom et al. (2010) previously reported that the same biodegradation rates will be obtained as long as the concentration ratio of the chemical compounds to bacterial cells is constant. However, the authors drew this conclusion based on experiments performed using a Pseudomonas graminis bacterial strain incubated in the presence of a single organic compound as the carbon source. At present, it is unclear whether this conclusion can be extrapolated to other bacteria species incubated in the presence of multiple organic compounds, and this warrants further study. Nevertheless, we made the same assumption (i.e., the same biodegradation rates will be obtained as long as the concentration ratio of the chemical compounds to bacterial cells is constant) as was done in previous studies that investigated the biodegradation of multiple organic compounds by different bacteria species (Vaïtilingom et al., 2011; Jaber et al., 2020, 2021). Hence, the bacteria concentration used was set to 106 cells per milliliter to maintain the same concentration ratio of the organic acids to bacterial cells. Table S2 shows the resulting concentrations of the organic and inorganic ions in the artificial cloud water used in these experiments.
During each experiment, aliquots of the solutions (0.6 mL) were taken every 2 h over 12 h. The organic acid concentrations in each filtered aliquot of solution were measured by ion chromatography (IC) using a Dionex ICS-1100 (Thermo Fisher Scientific Inc.) system. Details of the IC operation can be found in Sect. S4. To calculate the initial biodegradation rate, the time evolution of each organic acid concentration over 12 h was plotted and fitted with the following equation (Vaïtilingom et al., 2011; Jaber et al., 2020, 2021):
where k (s−1) is the rate constant obtained from the exponential fit to the decay of the organic acid. The following equation was used to calculate the biodegradation rate per bacteria cell (R):
where C0 (mol L−1) is the initial concentration of the organic acid, and [Cell]experiment (cells per liter) is the concentration of bacterial cells in the experiment. Control experiments were performed under illuminated and dark conditions using solutions that contained organic acids but no bacterial cells. The organic acids did not degrade in these control experiments.
3.1 Impact of pH on the survival and energetic metabolism of bacteria under illuminated and dark conditions
Figure 1 shows the survival rates and ADPATP ratios of the E. hormaechei B0910 and E. hormaechei pf0910 strains over time under illuminated and dark conditions at different artificial cloud water pH. The ADPATP ratio is used as an indicator of the bacteria's metabolic activity and survival rate in this study. Growing cells usually maintain a constant ADPATP ratio because, whenever there is a decrease in intracellular ATP production, its degradation product ADP will be resynthesized to form ATP to maintain intracellular ATP concentrations (Koutny et al., 2006; Guan and Liu, 2020). In contrast, when there is a disruption in the metabolism of ATP production, ATP cannot be resynthesized from ADP even though ATP is still converted to ADP, which will cause the ADPATP ratio to increase (Koutny et al., 2006; Guan and Liu, 2020).
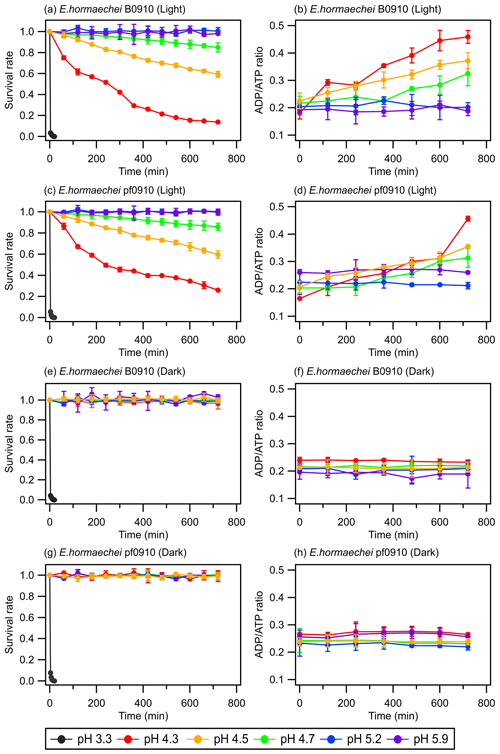
Figure 1Survival rates and ADPATP ratios of the E. hormaechei B0910 and E. hormaechei pf0910 strains at pH 3.3 to pH 5.9 under illuminated and dark conditions over time. The survival rate is defined as the number concentration of culturable viable cells divided by the initial number concentration of culturable viable cells at time point 0 min. Error bars represent 1 standard deviation from the mean of biological triplicates.
The artificial cloud water pH clearly had a significant effect on the survival rates and ADPATP ratios of the two strains. At pH 3.3, the concentrations of viable cells decreased to zero after 20 min, regardless of whether the strains were exposed to light. For pH 4.3, 4.5, and 4.7, the survival and ADPATP ratios of the two strains depended on whether they were exposed to light. There were no significant changes in the survival rates and ADPATP ratios for both strains under dark conditions. In contrast, the concentrations of viable cells for both strains gradually decreased when they were exposed to light. Consistent with the lower survival rates, the ADPATP ratios for both strains increased over time. The survival rates and ADPATP ratios were the lowest and highest, respectively, at pH 4.3 after 12 h of illumination. There were no significant changes in the survival rates and ADPATP ratios of both strains at pH 5.2 and 5.9 under illuminated and dark conditions.
Figure 1 clearly shows that the artificial cloud water pH and exposure to light can have a synergistic effect on the survival and energetic metabolism of E. hormaechei B0910 and E. hormaechei pf0910. Based on these results, both strains will likely survive during the daytime and nighttime in pH>5 cloud water. However, cloud water pH will play an important role in dictating the fraction of the bacteria that will survive in the daytime at pH 4 to 5. A low pH environment can lower the internal pH of cells, which affects the essential pH-dependent biological and cellular functions such as decreased enzymatic activity, compromised cellular processes (e.g., central metabolic pathways and ATP production), and protein denaturation in cells (Bearson et al., 1997; Lund et al., 2014). Our genomic analysis revealed that the two strains have genes encoding a F1F0-type ATP synthase, which can export protons from their cytoplasm to cope with pH stress (Krulwich et al., 2011). In addition, genes encoding potassium transporters, which may be involved in pH homeostasis (i.e., both Kup-type low-affinity and Kdp-type high-affinity potassium transporters; Brzoska et al., 2022) were found in the genome of both strains (Table S3). Our results indicated that both strains will likely survive in pH 4 to 5 cloud water at night. However, being in cloud water at pH 4 to 5 will likely negatively impact the ability of cells to tolerate sunlight, which will affect their survival during the daytime. Based on our results, we estimate that the half-lives of the bacteria strains in pH 4.3 cloud water under illumination conditions (e.g., light intensity and wavelengths) similar to those in our study are around 430 min. The half-lives of the bacteria strains in pH<4 are cloud water are lower. Based on our results, we estimate that the daytime and nighttime half-lives of the bacteria strains in pH 3.3 cloud water are around 2 min.
3.2 Compounds released by bacteria under acidic and illuminated conditions
Some bacteria species adapt to sunlight exposure and acidic environments by deploying adaptation strategies and defensive mechanisms such as undergoing DNA repair and aggregation-promoting and pigmentation mechanisms (Bearson et al., 1997; Davey and O'toole, 2000; Delort et al., 2010; Flemming and Wingender, 2010; Vaïtilingom et al., 2012; Matulova et al., 2014; Guan and Liu, 2020). Some of these adaptation strategies and defensive mechanisms will cause the bacteria to release organic compounds into cloud water (Davey and O'toole, 2000; Delort et al., 2010; Flemming and Wingender, 2010; Vaïtilingom et al., 2012; Matulova et al., 2014). In addition, bacterial cellular damage and lysis will lead to the release of biological material and organic compounds. To investigate the compounds released by E. hormaechei B0910 and E. hormaechei pf0910 during exposure to light and acidic environments, we used UPLC-MS to analyze the solutions in experiments where pH 4.3 and pH 5.9 artificial cloud water were used. The UPLC-MS measurements revealed that cell lysis led to the release of water-soluble and water-insoluble compounds when the two strains were exposed to light at pH 4.3. The quantities of these compounds changed with light exposure time. In contrast, no water-soluble and water-insoluble compounds were detected in the solutions of the two strains under dark conditions at pH 4.3 and under dark and illuminated conditions at pH 5.9. This suggested that these two strains did not release organic compounds, and the cells remained intact under these conditions. It is also possible that these two strains released organic compounds as an adaption strategy and/or defensive mechanism but that the concentrations of these compounds were below the detection limits of our UPLC-MS instrument.
Principal component analysis (PCA) with a 95 % confidence ellipse was applied to the UPLC-MS data of the detected water-soluble and water-insoluble compounds to identify discriminations between samples with different light exposure times. In each PCA plot (Fig. 2), samples with the same light exposure time clustered together. While there was a slight overlap between some of the clusters in the PCA plots, the clusters were mostly separated from one another. Partial least squares discrimination analysis (PLS-DA) was applied to the UPLC-MS data to identify water-soluble and water-insoluble compounds that showed significant changes in their relative abundances during exposure to light. A total of 259 water-soluble compounds and 215 water-insoluble compounds were identified for E. hormaechei B0910 (Fig. S3), while 209 water-soluble compounds and 251 water-insoluble compounds were identified for E. hormaechei pf0910 (Fig. S4). We identified the molecular formulas and chemical structures of 78 water-soluble compounds and 144 water-insoluble compounds released by E. hormaechei B0910 and 118 water-soluble compounds and 114 water-insoluble compounds released by E. hormaechei pf0910. These identified compounds were subsequently classified into different classes based on their chemical functionalities.
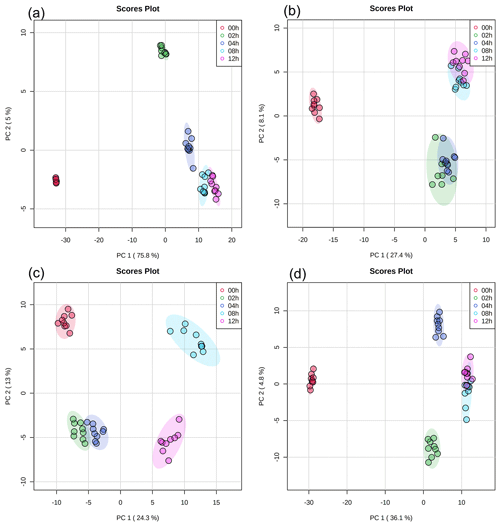
Figure 2PCA results of UPLC-MS data with (a) water-soluble compounds and (b) water-insoluble compounds from E. hormaechei B0910 and (c) water-soluble compounds and (d) water-insoluble compounds from E. hormaechei pf0910 during exposure to light at pH 4.3. Each cluster representing a different light exposure time (i.e., 0, 2, 4, 8, and 12 h) has nine points, since three samples were taken at each light exposure time, and UPLC-MS analysis was performed in triplicate for each sample.
Figures 3 and S5 show the time evolution of the UPLC-MS total ion chromatograph (TIC) signals of the different classes of water-soluble and water-insoluble compounds released by E. hormaechei B0910 and E. hormaechei pf0910 over time, respectively. The UPLC-MS TIC signals of the classes of water-soluble and water-insoluble compounds released by the two strains increased with light exposure time. The increase in the UPLC-MS TIC signals coincided with the decrease in the bacteria survival rate and the increase in the ADPATP ratio. Even though the heatmaps showed that some of the compounds had noticeable changes in their relative abundances during exposure to light (Figs. S3 and S4), the relative abundances of the different classes of compounds contributed to the total TIC at each time point did not change substantially (Figs. S6 and S7).
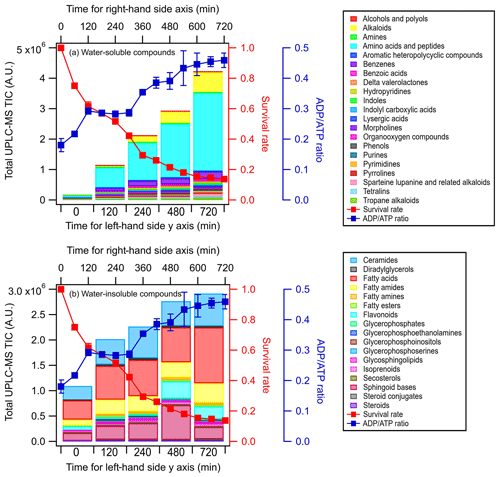
Figure 3Time evolution of the UPLC-MS total ion chromatograph (TIC) signals of (a) water-soluble compounds and (b) water-insoluble compounds from E. hormaechei B0910 during exposure to light at pH 4.3 over time. These compounds are classified based on their chemical functionality. Also shown is the time evolution of the survival rate and ADPATP ratio of E. hormaechei B0910.
To better understand the compounds released by the two strains, the OC and HC elemental ratios of the identified compounds were used to construct Van Krevelen (VK) diagrams. Regions of the VK diagrams were assigned to eight chemical classes based on the combined OC and HC ratios, namely lipids, unsaturated hydrocarbons, condensed aromatic structures, peptides, lignin, tannin, amino sugars, and carbohydrates (Table S4; Bianco et al., 2018; Laszakovits and Mackay, 2022). Rivas-Ubach et al. (2018) previously reported that the region of the VK diagram assigned to amino sugars overlaps with the region for nucleic acids. Figures S8 and S9 show the VK diagrams for water-soluble and water-insoluble compounds released by E. hormaechei B0910, respectively, while Figs. S10 and S11 show the VK diagrams for water-soluble and water-insoluble compounds released by E. hormaechei pf0910, respectively. The majority of the water-soluble and water-insoluble compounds released from both strains (50 % to 60 %) were assigned as lipids based on their OC and HC ratios, while the second most abundant compound class was peptides (10 % to 20 %). The two least abundant compound classes were amino sugars/nucleic acids and carbohydrates. Since the dry matter of a typical bacterial cell contains approximately 55 % proteins and amino acids, 24 % nucleic acids, 10 % carbohydrates, 7 % lipids, and 5 % inorganic minerals and trace elements (Watson et al., 2007), the differences in the abundance of compound classes detected vs. the dry matter of a typical bacterial cell indicated that cellular components were likely biologically and/or chemically modified during and after cell lysis during exposure to light. For instance, the large abundance of peptides detected could be a result of biological and/or chemical modifications of proteins and amino acids, which comprise majority of the dry matter of a typical bacterial cell. Peptide bonds are formed by biochemical reactions, where a water molecule is removed as the amino group of one amino acid is joined to the carboxyl group of a neighboring amino acid. The large abundance of lipids was unsurprising, since lipids are the main component of cell membranes, so large quantities of lipids are expected from the lysed cells. Most of the lipid molecules released during cell lysis may not have undergone biological and/or chemical modifications under our experimental conditions. The two least abundant compound classes were amino sugars/nucleic acids and carbohydrates. This was somewhat surprising since nucleic acids and carbohydrates are abundant in the dry matter of a typical bacterial cell. It is possible that these compounds were biologically and/or chemically modified to form other compounds (e.g., exopolymeric substances) during exposure to light (Matulova et al., 2014). In addition, the extraction procedure employed (Sect. S2) may not have extracted these compounds effectively for analysis. For instance, nucleic acids and carbohydrates are polar molecules, which are difficult to retain on the solid-phase extraction columns used in this study. These compounds may also have been poorly separated in UPLC and/or inefficiently ionized by ESI.
These detected compounds indicated that bacterial cell lysis could be a source for carbon in cloud water. Many of the compound classes detected in this study have previously been measured in atmospheric cloud water. For instance, large abundances of peptide-like compounds and lipid-like compounds have been measured in cloud water from Puy-de-Dôme (Bianco et al., 2018, 2019), which is consistent with the detection of large abundances of compounds assigned to the peptide and lipid compound classes in this study. This suggested that peptide-like and lipid-like compounds could be used as biomarkers to evaluate bacterial contributions to atmospheric samples. Previous studies have used fatty acids, which are integral building blocks of lipids, in atmospheric samples as biomarkers for characterizing and quantifying bacteria and assessing the atmospheric transport of bacteria (Kawamura et al., 2003; Lee et al., 2004; Tyagi et al., 2015). While this study shows that bacterial cell lysis will release large quantities of peptide-like and lipid-like compounds, using these compounds as biomarkers for bacterial cell lysis in atmospheric samples will likely be complex, as the concentrations of these compounds will likely change with time. This is because peptide-like and lipid-like compounds will undergo chemical and biological transformations after they have been released during cell lysis, which will impact their concentrations in atmospheric samples. Amino acids, which are building blocks of peptides, are known to undergo chemical reactions with oxidants in cloud water (Bianco et al., 2016). In addition, peptide-like and lipid-like compounds can be produced and/or consumed by cloud microorganisms to maintain their metabolism (Bianco et al., 2019; Jaber et al., 2021).
3.3 Impact of pH on the biodegradation of organic acids by bacteria under illuminated and dark conditions
The biodegradation of seven organic acids (i.e., formic acid, acetic acid, oxalic acid, maleic acid, malonic acid, glutaric acid, and MSA) that were mixed together were measured under dark and illuminated conditions at pH 4.3 and pH 5.9. Only some of the seven organic acids were biodegraded by the two strains. Based on our experimental conditions (liquid water content ≈1012 µg m−3; the density of water) and the Henry's law constants of the organic acids, these organic acids will be in the aqueous phase and are not expected to volatilize during these experiments. Thus, the observed decays were due to bacterial metabolism. E. hormaechei B0910 biodegraded formate and oxalate under dark and illuminated conditions at pH 4.3 and pH 5.9 and biodegraded malonate and maleate only under dark conditions at pH 4.3 and pH 5.9. In contrast, E. hormaechei pf0910 biodegraded only formate and oxalate under dark and illuminated conditions at pH 4.3 and pH 5.9. Biodegradation was not observed for acetate, MSA, and glutarate.
Table S5 summarizes the enzymes or metabolic pathways related to the biodegradation of organic acids in the two strains. Genes encoding formate dehydrogenases were identified in both genomes, which is consistent with the observed formate biodegradation. However, no known genes for oxalic acid biodegradation (Liu et al., 2021) were found in the genomes of both strains, which suggested the presence of yet-to-be-characterized pathways that catalyzed the biodegradation. Interestingly, a protein with a Cupin 2 domain was found in both genomes. The Cupin superfamily consists of a diverse range of enzymes including oxalate oxidase and oxalate decarboxylase that can biodegrade oxalic acid (Burrell et al., 2007).
Only the E. hormaechei B0910 strain was observed to biodegrade malonic acid. Interestingly, the malonyl-CoA-acyl carrier transcacylase observed in the E. hormaechei pf0910 strain seems to be a fusion protein, which may render it ineffective in utilizing malonic acid. Although no gene encoding maleate isomerase was identified in the genomes of both strains, the maleic acid biodegradation observed can be attributed to the activity of other enzymes with broad substrates specificity (Hatakeyama et al., 2000). The genes encoding for the small and large protein subunits that together form the 3-isopropylmalate dehydratase, the enzyme that isomerizes 2-isopropylmalate to 3-isopropylmalate, were found in both of the Enterobacter strains. The small and large protein subunits of this enzyme are homologous to the small (51 % amino acid identity) and large (59 % amino acid identity) protein subunit constituents of maleate hydratase (HbzIJ) from Pseudomonas alcaligenes NCIMB 9867 that converts maleate to D-malate (Liu et al., 2015). Given the high protein homology, we speculate that the 3-isopropylmalate dehydratase in the Enterobacter strains may have a broader substrate specificity than what is known, and it may be able to biodegrade maleate.
The lack of biodegradation of acetic acid, MSA, and glutaric acid in the experiments could be partly explained by the genomic information. Both strains have genes that encode enzymes involved in the biodegradation (Table S5) and associated uptake transporters (i.e., acetate permease (ActP) and succinate-acetate/proton symporter (SatP)) of acetic acid. The lack of the corresponding biodegradation in the experiments could be due to the low uptake of acetic acid by cells as ActP functions to scavenge low concentrations of the compound (Gimenez et al., 2003), while SatP could be inhibited by formic acid found in the cloud water medium (Sá-Pessoa et al., 2013). Genes encoding the two-component alkanesulfonate monooxygenase for MSA biodegradation were found in both strains, but they were likely not expressed, as sulfur was not deficient in the cloud water medium (Kahnert et al., 2000; Eichhorn and Leisinger, 2001), which is consistent with the absence of MSA biodegradation in the experiments. While genes encoding succinate-semialdehyde dehydrogenase/glutarate-semialdehyde dehydrogenase, which display a reversible conversion between glutarate-semialdehyde and glutarate in the KEGG database (Kanehisa et al., 2022), were found in both strains, to the best of our knowledge, there is no report of experimental results confirming that the reaction can go in the reverse direction from glutarate to glutarate-semialdehyde. In addition, a study of glutaric semialdehyde dehydrogenase reported on the irreversible nature of the catalysis of glutarate semialdehyde to glutarate (Ichihara and Ichihara, 1961). Thus, it is not surprising that glutarate biodegradation was not observed for the two strains.
Figure 4 summarizes the measured biodegradation rates of the organic acids for the two strains under dark and illuminated conditions at pH 4.3 and pH 5.9. These biodegradation rates were determined from fits to the decays of the organic acids from reaction time 0 to 12 h in each experiment (Sect. 2.4). The measured biodegradation rates were around 10−19 to 10−18 mol per cell per second, which were of the same order of magnitude as the bacterial strains isolated from cloud water and implemented into cloud models (Vaitilingom et al., 2010; Vaïtilingom et al., 2011; Fankhauser et al., 2019). Although both strains were affiliated to E. hormaechei, the artificial cloud water pH and exposure to light impacted their biodegradation of organic acids differently. The rates at which formate and oxalate were biodegraded by E. hormaechei B0910 had the following order: dark conditions at pH 5.9 > illuminated conditions at pH 5.9 > dark conditions at pH 4.3 > illuminated conditions at pH 4.3. This order was different for E. hormaechei pf0910, where dark conditions at pH 5.9 > dark conditions at pH 4.3 > illuminated conditions at pH 5.9 > illuminated conditions at pH 4.3. Despite the effects that the artificial cloud water pH and exposure to light had on the formate and oxalate biodegradation, the fastest and slowest biodegradation rates only differed by a factor of 1.4 to 3.7. Figure S12 compares the biodegradation rates measured at pH 4.3 vs. pH 5.9 and under illuminated vs. dark conditions. For the effect of artificial cloud water pH on the biodegradation of organic acids by E. hormaechei B0910, the differences in the biodegradation rates were statistically significant for the four acids (Student's t test; p value <0.05). Conversely, the differences in the biodegradation rates of formate and oxalate as a result of light exposure were statistically significant at pH 5.9 (Student's t test; p value <0.05). For the effect of artificial cloud water pH on the biodegradation of organic acids by E. hormaechei pf0910, only the difference in the dark biodegradation of oxalate was statistically significant (Student's t test; p value <0.05). In contrast, light exposure reduced the formate biodegradation rates significantly at both pH 4.3 and pH 5.9 (Student's t test; p value <0.05), and the oxalate biodegradation rate significantly at pH 5.9 (Student's t test; p value <0.05).
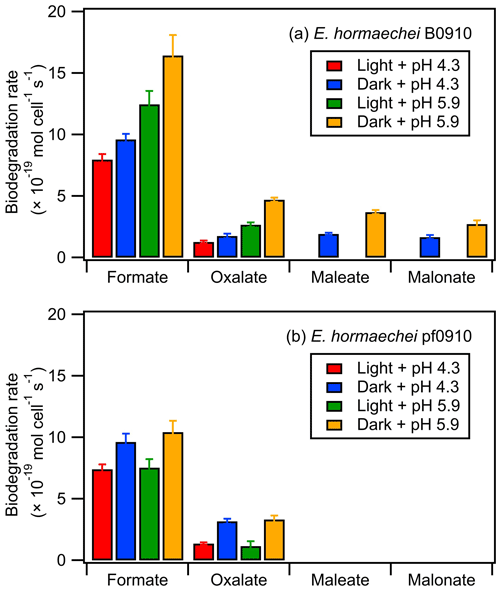
Figure 4Biodegradation rates of oxalate, maleate, and malonate by (a) E. hormaechei B0910 and (b) E. hormaechei pf0910 under light and dark conditions at pH 4.3 and pH 5.9. Error bars represent 1 standard deviation from the mean biodegradation rate.
The survival rates and ADPATP ratios of both strains were also monitored during the biodegradation experiments (Fig. S13). There were no significant changes in the survival rates and ADPATP ratios of both strains during the biodegradation process under dark conditions at pH 4.3 and under dark and illuminated conditions at pH 5.9. In contrast, the concentrations of viable cells gradually decreased until only 48 % and 60 % of the initial concentrations of viable cells remained at 12 h for E. hormaechei B0910 and E. hormaechei pf0910, respectively, during exposure to light at pH 4.3. The ADPATP ratios for both strains also increased during this time period, which is consistent with the lower metabolic activity and lower survival rate.
A simple kinetic analysis was performed to identify the factors that will impact the relative contributions of bacterial activity vs. chemistry in cloud water during the daytime and nighttime. Details of the calculations performed in this kinetic analysis can be found in Sect. S5. Our approach of considering daytime and nighttime processes separately was different from the approach used by previous studies, which determined the relative contributions of bacterial activity and chemical reactions on the degradation of organic compounds by only considering dark biodegradation processes and •OH photochemical reactions (Vaïtilingom et al., 2011; Jaber et al., 2020, 2021). Here, biodegradation rates that were measured under illuminated conditions were used for the daytime scenario, while biodegradation rates that were measured under dark conditions were used for the nighttime scenario. We used the average of biodegradation rates measured for the two strains for our calculations. Formate, oxalate, and malonate were chosen for our analysis, since their •OH and NO3• reaction rate constants were available in the literature. •OH and NO3• are the main tropospheric aqueous-phase free radicals during the daytime and nighttime, respectively (Herrmann et al., 2010). The average measured biodegradation rates of formate, oxalate, and malonate were first converted to biodegradation rate constants. These biodegradation rate constants and the corresponding •OH and NO3• reaction rate constants provided by the literature (Table 1) were subsequently used for calculations of the biodegradation rates and chemical reaction rates in cloud water (Sect. S5). A bacteria concentration of 8×107 cells per liter was assumed in our calculations for the daytime scenario at pH∼5 and the nighttime scenarios at pH∼4 and ∼5, which was the same bacteria concentration used in previous studies and represented the highest estimate of actual live bacteria concentrations (i.e., 100 % of metabolically active cells; Vaïtilingom et al., 2011; Jaber et al., 2020, 2021). Based on our investigations of the survival and energetic metabolism of bacteria under illuminated conditions at pH 4 to 5 (Fig. 1), we expect the bacteria concentrations to gradually decrease for the daytime scenario at pH∼4. Thus, for simplicity, we assumed a lower bacteria concentration in our calculations for the daytime scenario at pH∼4, whereby we multiplied the bacteria concentration of 8×107 cells per liter by a factor of 0.75. This factor was obtained by taking the average survival rates for the two strains from reaction time 0 to 12 h in our experiments conducted under illuminated conditions at pH 4.3 (Fig. S13). The rates of oxidation by •OH and NO3• chemical reactions will depend on their respective concentrations. Hence, we used the average •OH and NO3• concentrations reported by Herrmann et al. (2010) for remote, marine, and urban environments in our calculations (Table S6; Herrmann et al., 2010).
Table 1Rate constants used to estimate the loss rates by biodegradation and chemical reactions (i.e., •OH oxidation for daytime and NO3• for nighttime).
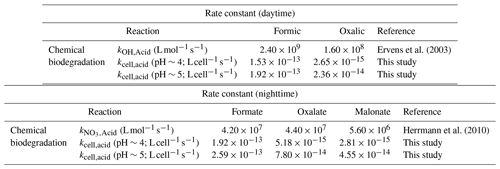
Calculations were performed for a variety of remote, marine, and urban environments with different formate, oxalate, and malonate concentrations that were previously reported in the literature (Table S7). Figure 5 shows the predicted relative contributions of bacterial activity vs. chemistry in remote, marine, and urban environments. •OH photochemistry will make a larger contribution to the daytime degradation of formate and oxalate in remote and marine environments due to the high •OH concentrations in these environments ( and M, respectively). In contrast, bacterial activity will play a bigger role in the daytime degradation of formate in urban environments due to their lower •OH concentrations ( M). However, •OH photochemistry will play a larger role in the daytime degradation of oxalate in urban environments due to the slow oxalate biodegradation rates. The low nighttime NO3• concentrations in remote and marine environments ( and M, respectively) will result in bacterial activity playing a bigger role in the nighttime degradation of formate, oxalate, and malonate in these two environments. In urban environments, bacterial activity will play a bigger role in the nighttime degradation of formate, but the nighttime degradation of oxalate and malonate will be dominated by NO3• chemistry due to the slow biodegradation rates of oxalate and malonate.
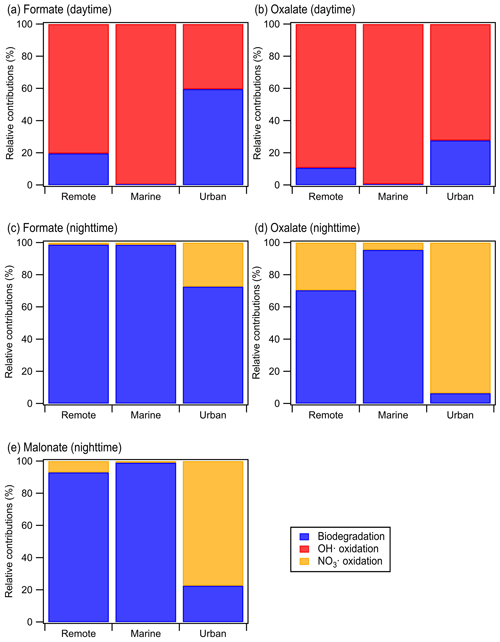
Figure 5Predicted relative contributions of bacterial activity and chemical reaction (i.e., •OH oxidation for daytime and NO3• for nighttime) to the degradation of organic compounds in the aqueous phase in remote, marine, and urban areas. This figure is based on estimated loss rates shown in Table S7.
Our simple kinetic analysis indicated that the organic acid, cloud water pH, radical oxidant concentration, and time of day (i.e., daytime vs. nighttime) will impact the relative contributions of bacterial activity vs. chemistry in the aqueous phase. However, there are a number of caveats that should be noted. First, the biodegradation rates used in this analysis were from experiments conducted at 25 ∘C, which may be more representative of warmer regions during the summer (e.g., Hong Kong and parts of South China). Slower biodegradation rates will likely be measured at lower temperatures (Ariya et al., 2002; Vaitilingom et al., 2010; Husárová et al., 2011; Vaïtilingom et al., 2011), which will impact the relative contributions of bacterial activity vs. chemistry. Second, our analysis did not account for how the presence of aqueous-phase oxidants (e.g., •OH in the daytime and NO3• in the nighttime) will impact the survival and energetic metabolism of bacteria, which in turn will impact the relative contributions of bacterial activity vs. chemistry. Third, our analysis did not account for the physical separation of cloud droplets containing bacteria cells from cell-free cloud droplets. Only a small fraction of cloud droplets will contain metabolically active bacteria cells, and the bacterial metabolism cannot affect the composition of organic acids in cell-free cloud droplets (Fankhauser et al., 2019; Khaled et al., 2021). Hence, only chemistry will govern the degradation of organic acids in cell-free droplets. Consequently, not accounting for the physical separation of cloud droplets containing bacteria cells from cell-free cloud droplets will result in an overestimation of the overall contribution of bacterial activity to the biodegradation of organic compounds (Fankhauser et al., 2019; Khaled et al., 2021). Fourth, our analysis only considers biodegradation and chemical reactions occurring in the aqueous phase and ignores gas-aqueous phase exchanges and gas-phase chemical reactions. Nah et al. (2018) previously showed that the gas–aqueous-phase partitioning of organic acids will depend on the Henry's law constant of the organic acid and acid dissociation constants, liquid water concentration, temperature, and pH (Sect. S6). Figure S14 shows that a significant fraction of formic acid will be in the gas phase at pH 4 and 5 under cloud water conditions, whereas all of oxalic acid, malonic acid, and maleic acid will be in the aqueous phase at pH 4 and 5 under cloud water conditions. This suggests that gas-phase chemical reactions will likely play an important role in consuming formic acid, whereas the consumption of oxalic acid, malonic acid, and maleic acid will likely mainly be through bacterial activity and chemical reactions in the aqueous phase. Quantifying the exact contributions of aqueous-phase bacterial activity vs. aqueous-phase chemistry vs. gas-phase chemistry under different cloud water pH conditions will require a multi-phase box model similar to the one used by Khaled et al. (2021). This is beyond the scope of the current study but can be a subject of future studies.
In this study, we investigated how cloud water pH and exposure to solar radiation impact the survival and energetic metabolism of bacteria and their ability to biodegrade organic acids in clouds. Laboratory experiments were performed using artificial solar radiation and artificial cloud water that mimicked the pH and composition of cloud water previously collected in South China, which is a region with fairly acidic cloud water (pH 3 to 5.9). Using two E. hormaechei strains that were isolated from ambient air in Hong Kong, we observed that the energetic metabolism and survival of both strains depended on the artificial cloud water pH. Low survival rates were observed for both strains at pH<4, regardless of whether the strains were exposed to light. At pH 4 to 5, the energetic metabolism and survival of both strains were only negatively impacted when they were exposed to light. In contrast, there were minimal effects on the energetic metabolism and survival of both strains when they were exposed to simulated sunlight at pH>5. In addition, the biodegradation of organic acids depended on the presence (or absence) of light and the artificial cloud water pH. The measured biodegradation rates were around 10−19 to 10−18 mol per cell per second, which were of the same order of magnitude as the bacterial strains isolated from cloud water and implemented into cloud models (Vaitilingom et al., 2010; Vaïtilingom et al., 2011; Fankhauser et al., 2019). Our analysis indicated that the organic acid, cloud water pH, radical oxidant concentration, and the time of day will impact the relative contributions of bacterial activity vs. chemistry in the aqueous phase.
This study has two important implications for our understanding of bacteria in clouds. First, this study underscores the importance of accounting for cloud water pH when simulating cloud processes involving metabolically active bacteria in atmospheric models, including microbiological–chemical interactions between live bacteria and organic matter. Results from this study imply the cloud water pH will impact the bacteria's ability to survive and thrive in during the daytime and/or nighttime. The pH of cloud water typically lies between 3 and 6 (Pye et al., 2020). Regions with high inputs of sulfuric acid and/or nitric acid combined with low inputs of ammonia, dust, and sea salt, such as South China, will have moderately acidic to highly acidic cloud water (Li et al., 2020; Pye et al., 2020; Shah et al., 2020; Qu and Han, 2021). Most of the bacteria in the atmosphere are neutrophiles that generally survive and thrive in less acidic environments. Hence, even though our study focuses on two Enterobacter strains, we hypothesize that cloud water pH will also affect the ability of other neutrophilic bacteria species to survive and remain metabolically active. Second, results from this study imply that it is important to consider the potential synergistic negative impacts that different stressors have on the survival and microbial activity of bacteria in clouds. Much of our current knowledge on the effect of different stressors (osmotic shocks, freeze–thaw cycles, and exposure to light and H2O2) on the survival of bacteria in clouds originate from a previous study by Joly et al. (2015), who investigated the impacts of these four stressors individually. However, as demonstrated in this study, when combined together, some stressors (in this case, cloud water pH and exposure to sunlight) can have synergistic negative impacts on the survival and microbial activity of bacteria in clouds.
While this study builds on our existing knowledge of how different stressors will impact the survival and energetic metabolism of bacteria and their ability to biodegrade organic matter in clouds, there are a number of caveats that should be noted. First, we were limited to using bacterial strains isolated from ambient air in this study due to the unavailability of bacteria isolates from cloud water in South China. Thus, if available, this work could be extended to bacteria isolates from cloud water in South China in the future to determine the pH conditions at which these isolates can survive and participate in microbiological–chemical interactions during the daytime and/or nighttime. The effect of cloud water pH on bacteria species that are reportedly common in cloud water (e.g., Sphingomonadales, Rhodospirillales, Rhizobiales, Burkholderiales, and Pseudomonadales; Vaïtilingom et al., 2012; Zhu et al., 2018; Peng et al., 2019) should also be investigated. Second, all the experiments in this study were conducted at 25 ∘C, which may be more representative of warmer regions during the summer (e.g., Hong Kong and parts of South China). Several studies have reported slower biodegradation rates at lower temperatures (Ariya et al., 2002; Vaitilingom et al., 2010; Husárová et al., 2011; Vaïtilingom et al., 2011), which suggests that cloud water temperature may influence the survival and energetic metabolism of bacteria. Third, the photon intensity in the photoreactor was kept constant in all the experiments. However, sunlight intensity will change throughout the day in the atmosphere. Fourth, this study does not consider how the presence of aqueous-phase oxidants (e.g., •OH in the daytime and NO3• in the nighttime) will impact the survival and energetic metabolism of bacteria in clouds. Hence, the effects of temperature, light intensity, and oxidants on the impact the survival and energetic metabolism of bacteria and their ability to biodegrade organic matter in clouds should be investigated in future studies.
The data used in this publication are available to the community and can either be accessed on request to the corresponding author (theodora.nah@cityu.edu.hk) or at https://doi.org/10.5281/zenodo.7045510 (Liu et al., 2022).
The supplement related to this article is available online at: https://doi.org/10.5194/acp-23-1731-2023-supplement.
YL, PKHL, and TN designed the study. YL conducted the experiments. YL, CKL, and ZS performed the data analysis. YL and TN wrote the paper, with contributions from all co-authors.
At least one of the (co-)authors is a member of the editorial board of Atmospheric Chemistry and Physics. The peer-review process was guided by an independent editor, and the authors also have no other competing interests to declare.
Publisher's note: Copernicus Publications remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
This work has been supported by the National Natural Science Foundation of China (project no. 42005081) and the Research Grants Council, University Grants Committee (project no. 11303720).
This paper was edited by Irena Grgić and reviewed by Pierre Amato and one anonymous referee.
Amato, P., Ménager, M., Sancelme, M., Laj, P., Mailhot, G., and Delort, A.-M.: Microbial population in cloud water at the Puy de Dôme: Implications for the chemistry of clouds, Atmos. Environ., 39, 4143–4153, https://doi.org/10.1016/j.atmosenv.2005.04.002, 2005.
Amato, P., Parazols, M., Sancelme, M., Mailhot, G., Laj, P., and Delort, A.-M.: An important oceanic source of micro-organisms for cloud water at the Puy de Dôme (France), Atmos. Environ., 41, 8253–8263, https://doi.org/10.1016/j.atmosenv.2007.06.022, 2007.
Amato, P., Joly, M., Besaury, L., Oudart, A., Taib, N., Mone, A. I., Deguillaume, L., Delort, A. M., and Debroas, D.: Active microorganisms thrive among extremely diverse communities in cloud water, PLoS One, 12, e0182869, https://doi.org/10.1371/journal.pone.0182869, 2017.
Amato, P., Besaury, L., Joly, M., Penaud, B., Deguillaume, L., and Delort, A.-M.: Metatranscriptomic exploration of microbial functioning in clouds, Sci. Rep.-UK, 9, 1–12, 2019.
Anglada, J. M., Martins-Costa, M., Francisco, J. S., and Ruiz-Lopez, M. F.: Interconnection of reactive oxygen species chemistry across the interfaces of atmospheric, environmental, and biological processes, Accounts Chem. Res., 48, 575–583, https://doi.org/10.1021/ar500412p, 2015.
Ariya, P. A., Nepotchatykh, O., Ignatova, O., and Amyot, M.: Microbiological degradation of atmospheric organic compounds, Geophys. Res. Lett., 29, 34-31–34-34, https://doi.org/10.1029/2002gl015637, 2002.
Attard, E., Yang, H., Delort, A.-M., Amato, P., Pöschl, U., Glaux, C., Koop, T., and Morris, C. E.: Effects of atmospheric conditions on ice nucleation activity of Pseudomonas, Atmos. Chem. Phys., 12, 10667–10677, https://doi.org/10.5194/acp-12-10667-2012, 2012.
Bauer, H., Kasper-Giebl, A., Loflund, M., Giebl, H., Hitzenberger, R., Zibuschka, F., and Puxbaum, H.: The contribution of bacteria and fungal spores to the organic carbon content of cloud water, precipitation and aerosols, Atmos. Res., 64, 109–119, https://doi.org/10.1016/s0169-8095(02)00084-4, 2002.
Bearson, S., Bearson, B., and Foster, J. W.: Acid stress responses in enterobacteria, FEMS Microbiol. Lett., 147, 173–180, https://doi.org/10.1111/j.1574-6968.1997.tb10238.x, 1997.
Bianco, A., Voyard, G., Deguillaume, L., Mailhot, G., and Brigante, M.: Improving the characterization of dissolved organic carbon in cloud water: Amino acids and their impact on the oxidant capacity, Sci. Rep.-UK, 6, 37420, https://doi.org/10.1038/srep37420, 2016.
Bianco, A., Deguillaume, L., Vaitilingom, M., Nicol, E., Baray, J. L., Chaumerliac, N., and Bridoux, M.: Molecular Characterization of Cloud Water Samples Collected at the Puy de Dome (France) by Fourier Transform Ion Cyclotron Resonance Mass Spectrometry, Environ. Sci. Technol., 52, 10275–10285, https://doi.org/10.1021/acs.est.8b01964, 2018.
Bianco, A., Deguillaume, L., Chaumerliac, N., Vaïtilingom, M., Wang, M., Delort, A.-M., and Bridoux, M. C.: Effect of endogenous microbiota on the molecular composition of cloud water: a study by Fourier-transform ion cyclotron resonance mass spectrometry (FT-ICR MS), Sci. Rep.-UK, 9, 7663, https://doi.org/10.1038/s41598-019-44149-8, 2019.
Brzoska, R. M., Edelmann, R. E., and Bollmann, A.: Physiological and Genomic Characterization of Two Novel Bacteroidota Strains Asinibacterium spp. OR43 and OR53, Bacteria, 1, 33–47, 2022.
Burrell, M. R., Just, V. J., Bowater, L., Fairhurst, S. A., Requena, L., Lawson, D. M., and Bornemann, S.: Oxalate decarboxylase and oxalate oxidase activities can be interchanged with a specificity switch of up to 282 000 by mutating an active site lid, Biochemistry, 46, 12327–12336, https://doi.org/10.1021/bi700947s, 2007.
Burrows, S. M., Elbert, W., Lawrence, M. G., and Pöschl, U.: Bacteria in the global atmosphere – Part 1: Review and synthesis of literature data for different ecosystems, Atmos. Chem. Phys., 9, 9263–9280, https://doi.org/10.5194/acp-9-9263-2009, 2009.
Chen, X., Ran, P., Ho, K., Lu, W., Li, B., Gu, Z., Song, C., and Wang, J.: Concentrations and Size Distributions of Airborne Microorganisms in Guangzhou during Summer, Aerosol Air Qual. Res., 12, 1336–1344, https://doi.org/10.4209/aaqr.2012.03.0066, 2012.
Davey, M. E. and O'toole, G. A.: Microbial biofilms: from ecology to molecular genetics, Microbiol. Mol. Biol. R., 64, 847–867, https://doi.org/10.1128/MMBR.64.4.847-867.2000, 2000.
Delort, A.-M., Vaïtilingom, M., Amato, P., Sancelme, M., Parazols, M., Mailhot, G., Laj, P., and Deguillaume, L.: A short overview of the microbial population in clouds: Potential roles in atmospheric chemistry and nucleation processes, Atmos. Res., 98, 249–260, https://doi.org/10.1016/j.atmosres.2010.07.004, 2010.
Després, V., Huffman, J. A., Burrows, S. M., Hoose, C., Safatov, A., Buryak, G., Fröhlich-Nowoisky, J., Elbert, W., Andreae, M., Pöschl, U., and Jaenicke, R.: Primary biological aerosol particles in the atmosphere: a review, Tellus B, 64, 15598, https://doi.org/10.3402/tellusb.v64i0.15598, 2012.
Ding, W., Li, L., Han, Y., Liu, J., and Liu, J.: Site-related and seasonal variation of bioaerosol emission in an indoor wastewater treatment station: level, characteristics of particle size, and microbial structure, Aerobiologia, 32, 211–224, https://doi.org/10.1007/s10453-015-9391-5, 2015.
Eichhorn, E. and Leisinger, T.: Escherichia coli utilizes methanesulfonate and L-cysteate as sole sulfur sources for growth, FEMS Microbiol. Lett., 205, 271–275, https://doi.org/10.1111/j.1574-6968.2001.tb10960.x, 2001.
Ervens, B. and Amato, P.: The global impact of bacterial processes on carbon mass, Atmos. Chem. Phys., 20, 1777–1794, https://doi.org/10.5194/acp-20-1777-2020, 2020.
Ervens, B., Gligorovski, S., and Herrmann, H.: Temperature-dependent rate constants for hydroxyl radical reactions with organic compounds in aqueous solutions, Phys. Chem. Chem. Phys., 5, 1811–1824, https://doi.org/10.1039/B300072A, 2003.
Fankhauser, A. M., Antonio, D. D., Krell, A., Alston, S. J., Banta, S., and McNeill, V. F.: Constraining the Impact of Bacteria on the Aqueous Atmospheric Chemistry of Small Organic Compounds, ACS Earth and Space Chemistry, 3, 1485–1491, https://doi.org/10.1021/acsearthspacechem.9b00054, 2019.
Flemming, H. C. and Wingender, J.: The biofilm matrix, Nat. Rev. Microbiol., 8, 623–633, https://doi.org/10.1038/nrmicro2415, 2010.
George, K. M., Ruthenburg, T. C., Smith, J., Yu, L., Zhang, Q., Anastasio, C., and Dillner, A. M.: FT-IR quantification of the carbonyl functional group in aqueous-phase secondary organic aerosol from phenols, Atmos. Environ., 100, 230–237, https://doi.org/10.1016/j.atmosenv.2014.11.011, 2015.
Gimenez, R., Nuñez, M. F., Badia, J., Aguilar, J., and Baldoma, L.: The gene yjcG, cotranscribed with the gene acs, encodes an acetate permease in Escherichia coli, J. Bacteriol., 185, 6448–6455, https://doi.org/10.1128/JB.185.21.6448-6455.2003, 2003.
Guan, N. and Liu, L.: Microbial response to acid stress: mechanisms and applications, Appl. Microbiol. Biot., 104, 51–65, https://doi.org/10.1007/s00253-019-10226-1, 2020.
Hatakeyama, K., Goto, M., Kobayashi, M., Terasawa, M., and Yukawa, H.: Analysis of oxidation sensitivity of maleate cis-trans isomerase from Serratia marcescens, Biosci. Biotech. Bioch., 64, 1477–1485, https://doi.org/10.1271/bbb.64.1477, 2000.
Herrmann, H., Hoffmann, D., Schaefer, T., Brauer, P., and Tilgner, A.: Tropospheric aqueous-phase free-radical chemistry: radical sources, spectra, reaction kinetics and prediction tools, ChemPhysChem, 11, 3796–3822, https://doi.org/10.1002/cphc.201000533, 2010.
Hu, W., Niu, H. Y., Murata, K., Wu, Z. J., Hu, M., Kojima, T., and Zhang, D. Z.: Bacteria in atmospheric waters: Detection, characteristics and implications, Atmos. Environ., 179, 201–221, https://doi.org/10.1016/j.atmosenv.2018.02.026, 2018.
Huang, D. D., Zhang, Q., Cheung, H. H. Y., Yu, L., Zhou, S., Anastasio, C., Smith, J. D., and Chan, C. K.: Formation and Evolution of aqSOA from Aqueous-Phase Reactions of Phenolic Carbonyls: Comparison between Ammonium Sulfate and Ammonium Nitrate Solutions, Environ. Sci. Technol., 52, 9215–9224, https://doi.org/10.1021/acs.est.8b03441, 2018.
Huang, S., Hu, W., Chen, J., Wu, Z., Zhang, D., and Fu, P.: Overview of biological ice nucleating particles in the atmosphere, Environ. Int., 146, 106197, https://doi.org/10.1016/j.envint.2020.106197, 2021.
Husárová, S., Vaïtilingom, M., Deguillaume, L., Traikia, M., Vinatier, V., Sancelme, M., Amato, P., Matulová, M., and Delort, A.-M.: Biotransformation of methanol and formaldehyde by bacteria isolated from clouds. Comparison with radical chemistry, Atmos. Environ., 45, 6093–6102, https://doi.org/10.1016/j.atmosenv.2011.06.035, 2011.
Ichihara, A. and Ichihara, E. A.: Metabolism of L-Lysine by Bacterial Enzymes V. Glutaric Semialdehyde Dehydrogenase, J. Biochem., 49, 154–157, https://doi.org/10.1093/oxfordjournals.jbchem.a127272, 1961.
Jaber, S., Lallement, A., Sancelme, M., Leremboure, M., Mailhot, G., Ervens, B., and Delort, A.-M.: Biodegradation of phenol and catechol in cloud water: comparison to chemical oxidation in the atmospheric multiphase system, Atmos. Chem. Phys., 20, 4987–4997, https://doi.org/10.5194/acp-20-4987-2020, 2020.
Jaber, S., Joly, M., Brissy, M., Leremboure, M., Khaled, A., Ervens, B., and Delort, A.-M.: Biotic and abiotic transformation of amino acids in cloud water: experimental studies and atmospheric implications, Biogeosciences, 18, 1067–1080, https://doi.org/10.5194/bg-18-1067-2021, 2021.
Jaenicke, R.: Abundance of cellular material and proteins in the atmosphere, Science, 308, 73, https://doi.org/10.1126/science.1106335, 2005.
Joly, M., Amato, P., Sancelme, M., Vinatier, V., Abrantes, M., Deguillaume, L., and Delort, A.-M.: Survival of microbial isolates from clouds toward simulated atmospheric stress factors, Atmos. Environ., 117, 92–98, https://doi.org/10.1016/j.atmosenv.2015.07.009, 2015.
Kahnert, A., Vermeij, P., Wietek, C., James, P., Leisinger, T., and Kertesz, M. A.: The ssu locus plays a key role in organosulfur metabolism in Pseudomonas putida S-313, J. Bacteriol., 182, 2869–2878, https://doi.org/10.1128/JB.182.10.2869-2878.2000, 2000.
Kanehisa, M., Sato, Y., and Kawashima, M.: KEGG mapping tools for uncovering hidden features in biological data, Protein Sci., 31, 47–53, https://doi.org/10.1002/pro.4172, 2022.
Kawamura, K., Ishimura, Y., and Yamazaki, K.: Four years' observations of terrestrial lipid class compounds in marine aerosols from the western North Pacific, Global Biogeochem. Cy., 17, 1003, https://doi.org/10.1029/2001gb001810, 2003.
Khaled, A., Zhang, M., Amato, P., Delort, A.-M., and Ervens, B.: Biodegradation by bacteria in clouds: an underestimated sink for some organics in the atmospheric multiphase system, Atmos. Chem. Phys., 21, 3123–3141, https://doi.org/10.5194/acp-21-3123-2021, 2021.
Koutny, M., Sancelme, M., Dabin, C., Pichon, N., Delort, A.-M., and Lemaire, J.: Acquired biodegradability of polyethylenes containing pro-oxidant additives, Polym. Degrad. Stabil., 91, 1495–1503, https://doi.org/10.1016/j.polymdegradstab.2005.10.007, 2006.
Krulwich, T. A., Sachs, G., and Padan, E.: Molecular aspects of bacterial pH sensing and homeostasis, Nat. Rev. Microbiol., 9, 330–343, 2011.
Krumins, V., Mainelis, G., Kerkhof, L. J., and Fennell, D. E.: Substrate-dependent rRNA production in an airborne bacterium, Environ. Sci. Tech. Let., 1, 376–381, 2014.
Laszakovits, J. R. and MacKay, A. A.: Data-Based Chemical Class Regions for Van Krevelen Diagrams, J. Am. Soc. Mass Spectr., 33, 198–202, https://doi.org/10.1021/jasms.1c00230, 2022.
Lee, A. K. Y., Chan, C. K., Fang, M., and Lau, A. P. S.: The 3-hydroxy fatty acids as biomarkers for quantification and characterization of endotoxins and Gram-negative bacteria in atmospheric aerosols in Hong Kong, Atmos. Environ., 38, 6307–6317, https://doi.org/10.1016/j.atmosenv.2004.08.013, 2004.
Li, T., Wang, Z., Wang, Y., Wu, C., Liang, Y., Xia, M., Yu, C., Yun, H., Wang, W., Wang, Y., Guo, J., Herrmann, H., and Wang, T.: Chemical characteristics of cloud water and the impacts on aerosol properties at a subtropical mountain site in Hong Kong SAR, Atmos. Chem. Phys., 20, 391–407, https://doi.org/10.5194/acp-20-391-2020, 2020.
Li, Y., He, Y., Lam, C. H., and Nah, T.: Environmental photochemistry of organic UV filter butyl methoxydibenzoylmethane: Implications for photochemical fate in surface waters, Sci. Total Environ., 839, 156145, https://doi.org/10.1016/j.scitotenv.2022.156145, 2022.
Liu, K., Xu, Y., and Zhou, N.-Y.: Identification of a specific maleate hydratase in the direct hydrolysis route of the gentisate pathway, Appl. Environ. Microb., 81, 5753–5760, https://doi.org/10.1128/AEM.00975-15, 2015.
Liu, M., Devlin, J. C., Hu, J., Volkova, A., Battaglia, T. W., Ho, M., Asplin, J. R., Byrd, A., Li, H., and Ruggles, K. V.: Microbial genetic and transcriptional contributions to oxalate degradation by the gut microbiota in health and disease, Elife, 10, e63642, https://doi.org/10.7554/eLife.63642, 2021.
Liu, Y., Lim, C. K., Shen, Z., Lee, P. K. H., and Nah, T.: Effects of pH and light exposure on the survival of bacteria and their ability to biodegrade organic compounds in clouds: Implications for microbial activity in acidic cloud water, Zenodo [data set], https://doi.org/10.5281/zenodo.7045510, 2022.
Löflund, M., Kasper-Giebl, A., Schuster, B., Giebl, H., Hitzenberger, R., and Puxbaum, H.: Formic, acetic, oxalic, malonic and succinic acid concentrations and their contribution to organic carbon in cloud water, Atmos. Environ., 36, 1553–1558, https://doi.org/10.1016/S1352-2310(01)00573-8, 2002.
Lund, P., Tramonti, A., and De Biase, D.: Coping with low pH: molecular strategies in neutralophilic bacteria, FEMS Microbiol. Rev., 38, 1091–1125, https://doi.org/10.1111/1574-6976.12076, 2014.
Matulova, M., Husarova, S., Capek, P., Sancelme, M., and Delort, A. M.: Biotransformation of various saccharides and production of exopolymeric substances by cloud-borne Bacillus sp. 3B6, Environ. Sci. Technol., 48, 14238–14247, https://doi.org/10.1021/es501350s, 2014.
Misovich, M. V., Hettiyadura, A. P. S., Jiang, W. Q., Zhang, Q., and Laskin, A.: Molecular-Level Study of the Photo-Oxidation of Aqueous-Phase Guaiacyl Acetone in the Presence of C-3*: Formation of Brown Carbon Products, ACS Earth and Space Chemistry, 5, 1983–1996, https://doi.org/10.1021/acsearthspacechem.1c00103, 2021.
Möhler, O., DeMott, P. J., Vali, G., and Levin, Z.: Microbiology and atmospheric processes: the role of biological particles in cloud physics, Biogeosciences, 4, 1059–1071, https://doi.org/10.5194/bg-4-1059-2007, 2007.
Morris, C. E., Conen, F., Huffman, J. A., Phillips, V., Pöschl, U., and Sands, D. C.: Bioprecipitation: a feedback cycle linking earth history, ecosystem dynamics and land use through biological ice nucleators in the atmosphere, Glob. Change Biol., 20, 341–351, https://doi.org/10.1111/gcb.12447, 2014.
Morris, C. E., Soubeyrand, S., Bigg, E. K., Creamean, J. M., and Sands, D. C.: Mapping Rainfall Feedback to Reveal the Potential Sensitivity of Precipitation to Biological Aerosols, B. Am. Meteorol. Soc., 98, 1109–1118, https://doi.org/10.1175/BAMS-D-15-00293.1, 2017.
Nah, T., Guo, H., Sullivan, A. P., Chen, Y., Tanner, D. J., Nenes, A., Russell, A., Ng, N. L., Huey, L. G., and Weber, R. J.: Characterization of aerosol composition, aerosol acidity, and organic acid partitioning at an agriculturally intensive rural southeastern US site, Atmos. Chem. Phys., 18, 11471–11491, https://doi.org/10.5194/acp-18-11471-2018, 2018.
Péguilhan, R., Besaury, L., Rossi, F., Enault, F., Baray, J.-L., Deguillaume, L., and Amato, P.: Rainfalls sprinkle cloud bacterial diversity while scavenging biomass, FEMS Microbiol. Ecol., 97, fiab144, https://doi.org/10.1093/femsec/fiab144, 2021.
Peng, J., Zhou, S., Xiao, K., Zeng, J., Yao, C., Lu, S., Zhang, W., Fu, Y., Yang, Y., and Bi, X.: Diversity of bacteria in cloud water collected at a National Atmospheric Monitoring Station in Southern China, Atmos. Res., 218, 176–182, https://doi.org/10.1016/j.atmosres.2018.12.004, 2019.
Prokof'eva, T. V., Shoba, S. A., Lysak, L. V., Ivanova, A. E., Glushakova, A. M., Shishkov, V. A., Lapygina, E. V., Shilaika, P. D., and Glebova, A. A.: Organic Constituents and Biota in the Urban Atmospheric Solid Aerosol: Potential Effects on Urban Soils, Eurasian Soil Sci.+, 54, 1532–1545, https://doi.org/10.1134/S1064229321100094, 2021.
Pye, H. O. T., Nenes, A., Alexander, B., Ault, A. P., Barth, M. C., Clegg, S. L., Collett Jr., J. L., Fahey, K. M., Hennigan, C. J., Herrmann, H., Kanakidou, M., Kelly, J. T., Ku, I.-T., McNeill, V. F., Riemer, N., Schaefer, T., Shi, G., Tilgner, A., Walker, J. T., Wang, T., Weber, R., Xing, J., Zaveri, R. A., and Zuend, A.: The acidity of atmospheric particles and clouds, Atmos. Chem. Phys., 20, 4809–4888, https://doi.org/10.5194/acp-20-4809-2020, 2020.
Qu, R. and Han, G.: A critical review of the variation in rainwater acidity in 24 Chinese cities during 1982–2018, Elementa: Science of the Anthropocene, 9, 00142, https://doi.org/10.1525/elementa.2021.00142, 2021.
Rivas-Ubach, A., Liu, Y., Bianchi, T. S., Tolic, N., Jansson, C., and Pasa-Tolic, L.: Moving beyond the van Krevelen diagram: A new stoichiometric approach for compound classification in organisms, Anal. Chem., 90, 6152–6160, 2018.
Romano, S., Di Salvo, M., Rispoli, G., Alifano, P., Perrone, M. R., and Tala, A.: Airborne bacteria in the Central Mediterranean: Structure and role of meteorology and air mass transport, Sci. Total Environ., 697, 134020, https://doi.org/10.1016/j.scitotenv.2019.134020, 2019.
Romano, S., Fragola, M., Alifano, P., Perrone, M. R., and Talà, A.: Potential Human and Plant Pathogenic Species in Airborne PM10 Samples and Relationships with Chemical Components and Meteorological Parameters, Atmosphere, 12, 654, https://doi.org/10.3390/atmos12050654, 2021.
Ruiz-Gil, T., Acuña, J. J., Fujiyoshi, S., Tanaka, D., Noda, J., Maruyama, F., and Jorquera, M. A.: Airborne bacterial communities of outdoor environments and their associated influencing factors, Environ. Int., 145, 106156, https://doi.org/10.1016/j.envint.2020.106156, 2020.
Sá-Pessoa, J., Paiva, S., Ribas, D., Silva, I. J., Viegas, S. C., Arraiano, C. M., and Casal, M.: SATP (YaaH), a succinate–acetate transporter protein in Escherichia coli, Biochem. J., 454, 585–595, https://doi.org/10.1042/BJ20130412, 2013.
Shah, V., Jacob, D. J., Moch, J. M., Wang, X., and Zhai, S.: Global modeling of cloud water acidity, precipitation acidity, and acid inputs to ecosystems, Atmos. Chem. Phys., 20, 12223–12245, https://doi.org/10.5194/acp-20-12223-2020, 2020.
Sun, X., Wang, Y., Li, H., Yang, X., Sun, L., Wang, X., Wang, T., and Wang, W.: Organic acids in cloud water and rainwater at a mountain site in acid rain areas of South China, Environ. Sci. Pollut. R., 23, 9529–9539, https://doi.org/10.1007/s11356-016-6038-1, 2016.
Tsai, Y. I. and Kuo, S.-C.: Contributions of low molecular weight carboxylic acids to aerosols and wet deposition in a natural subtropical broad-leaved forest environment, Atmos. Environ., 81, 270–279, https://doi.org/10.1016/j.atmosenv.2013.08.061, 2013.
Tyagi, P., Ishimura, Y., and Kawamura, K.: Hydroxy fatty acids in marine aerosols as microbial tracers: 4-year study on β- and ω-hydroxy fatty acids from remote Chichijima Island in the western North Pacific, Atmos. Environ., 115, 89–100, 2015.
Vaitilingom, M., Amato, P., Sancelme, M., Laj, P., Leriche, M., and Delort, A. M.: Contribution of microbial activity to carbon chemistry in clouds, Appl. Environ. Microb., 76, 23–29, https://doi.org/10.1128/AEM.01127-09, 2010.
Vaïtilingom, M., Charbouillot, T., Deguillaume, L., Maisonobe, R., Parazols, M., Amato, P., Sancelme, M., and Delort, A.-M.: Atmospheric chemistry of carboxylic acids: microbial implication versus photochemistry, Atmos. Chem. Phys., 11, 8721–8733, https://doi.org/10.5194/acp-11-8721-2011, 2011.
Vaïtilingom, M., Attard, E., Gaiani, N., Sancelme, M., Deguillaume, L., Flossmann, A. I., Amato, P., and Delort, A.-M.: Long-term features of cloud microbiology at the puy de Dôme (France), Atmos. Environ., 56, 88–100, https://doi.org/10.1016/j.atmosenv.2012.03.072, 2012.
Vaitilingom, M., Deguillaume, L., Vinatier, V., Sancelme, M., Amato, P., Chaumerliac, N., and Delort, A. M.: Potential impact of microbial activity on the oxidant capacity and organic carbon budget in clouds, P. Natl. Acad. Sci. USA, 110, 559–564, https://doi.org/10.1073/pnas.1205743110, 2013.
Watson, J., Baker, T., and Bell, S.: Molecular biology of the gene, 6th edn., Cold Spring Harbor Laboratory Press, ISBN-10 080539592X, ISBN-13 978-0805395921, 2007.
Wei, M., Xu, C., Chen, J., Zhu, C., Li, J., and Lv, G.: Characteristics of bacterial community in cloud water at Mt Tai: similarity and disparity under polluted and non-polluted cloud episodes, Atmos. Chem. Phys., 17, 5253–5270, https://doi.org/10.5194/acp-17-5253-2017, 2017.
Zhang, M., Khaled, A., Amato, P., Delort, A.-M., and Ervens, B.: Sensitivities to biological aerosol particle properties and ageing processes: potential implications for aerosol–cloud interactions and optical properties, Atmos. Chem. Phys., 21, 3699–3724, https://doi.org/10.5194/acp-21-3699-2021, 2021.
Zhou, H., Wang, X., Li, Z., Kuang, Y., Mao, D., and Luo, Y.: Occurrence and Distribution of Urban Dust-Associated Bacterial Antibiotic Resistance in Northern China, Environ. Sci. Tech. Let., 5, 50–55, https://doi.org/10.1021/acs.estlett.7b00571, 2018.
Zhu, C., Chen, J., Wang, X., Li, J., Wei, M., Xu, C., Xu, X., Ding, A., and Collett, J. L.: Chemical Composition and Bacterial Community in Size-Resolved Cloud Water at the Summit of Mt. Tai, China, Aerosol Air Qual. Res., 18, 1–14, https://doi.org/10.4209/aaqr.2016.11.0493, 2018.





